- Record: found
- Abstract: found
- Article: found
Quantitative Characterization of Cellular Membrane-Receptor Heterogeneity through Statistical and Computational Modeling
Read this article at
Abstract
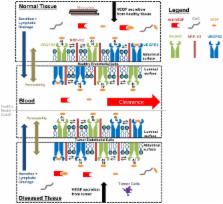
Cell population heterogeneity can affect cellular response and is a major factor in drug resistance. However, there are few techniques available to represent and explore how heterogeneity is linked to population response. Recent high-throughput genomic, proteomic, and cellomic approaches offer opportunities for profiling heterogeneity on several scales. We have recently examined heterogeneity in vascular endothelial growth factor receptor (VEGFR) membrane localization in endothelial cells. We and others processed the heterogeneous data through ensemble averaging and integrated the data into computational models of anti-angiogenic drug effects in breast cancer. Here we show that additional modeling insight can be gained when cellular heterogeneity is considered. We present comprehensive statistical and computational methods for analyzing cellomic data sets and integrating them into deterministic models. We present a novel method for optimizing the fit of statistical distributions to heterogeneous data sets to preserve important data and exclude outliers. We compare methods of representing heterogeneous data and show methodology can affect model predictions up to 3.9-fold. We find that VEGF levels, a target for tuning angiogenesis, are more sensitive to VEGFR1 cell surface levels than VEGFR2; updating VEGFR1 levels in the tumor model gave a 64% change in free VEGF levels in the blood compartment, whereas updating VEGFR2 levels gave a 17% change. Furthermore, we find that subpopulations of tumor cells and tumor endothelial cells (tEC) expressing high levels of VEGFR (>35,000 VEGFR/cell) negate anti-VEGF treatments. We show that lowering the VEGFR membrane insertion rate for these subpopulations recovers the anti-angiogenic effect of anti-VEGF treatment, revealing new treatment targets for specific tumor cell subpopulations. This novel method of characterizing heterogeneous distributions shows for the first time how different representations of the same data set lead to different predictions of drug efficacy.
Related collections
Most cited references49
- Record: found
- Abstract: found
- Article: not found
Role of the vascular endothelial growth factor pathway in tumor growth and angiogenesis.
- Record: found
- Abstract: found
- Article: not found
Vascular endothelial growth factor (VEGF) stimulates neurogenesis in vitro and in vivo.
- Record: found
- Abstract: found
- Article: not found