- Record: found
- Abstract: found
- Article: found
A novel deletion mutation in KMT2A identified in a child with ID/DD and blood eosinophilia
Read this article at
Abstract
Background
The KMT2A gene encoded lysine methyltransferase plays an essential role in regulating gene expression during early development and hematopoiesis. To date, 92 different mutations of KMT2A have been curated in the human gene mutation database (HGMD), resulting in Wiedemann-Steiner syndrome (WDSTS) and intellectual disability (ID)/developmental delay (DD).
Case presentation
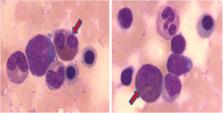
In this report, we present a de novo heterozygous deletion mutation [c.74delG; p. (Gly26Alafs*2)] in the KMT2A gene, which was identified by trio-based whole exome sequencing from a 5.5-year-old boy with ID/DD, stereotypic hand movements and blood eosinophilia. Many deleterious germline mutations of KMT2A have been documented to affect development of central nervous system, oral and craniofacial tissues, but not blood eosinophils.
Conclusions
This is the first report of a rare case with ID/DD as well as eosinophilia, resulting from a previously undescribed null mutation of KMT2A. Our findings expand the phenotypical spectrum in affected individuals with KMT2A mutations, and may shed some insight into the role of KMT2A in eosinophil metabolism.
Related collections
Most cited references22
- Record: found
- Abstract: found
- Article: not found
Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability.
- Record: found
- Abstract: found
- Article: not found
Genes with de novo mutations are shared by four neuropsychiatric disorders discovered from NPdenovo database.
- Record: found
- Abstract: found
- Article: not found