- Record: found
- Abstract: found
- Article: found
Bayesian methods outperform parsimony but at the expense of precision in the estimation of phylogeny from discrete morphological data
Read this article at
Abstract
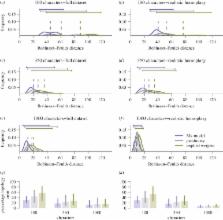
Different analytical methods can yield competing interpretations of evolutionary history and, currently, there is no definitive method for phylogenetic reconstruction using morphological data. Parsimony has been the primary method for analysing morphological data, but there has been a resurgence of interest in the likelihood-based Mk-model. Here, we test the performance of the Bayesian implementation of the Mk-model relative to both equal and implied-weight implementations of parsimony. Using simulated morphological data, we demonstrate that the Mk-model outperforms equal-weights parsimony in terms of topological accuracy, and implied-weights performs the most poorly. However, the Mk-model produces phylogenies that have less resolution than parsimony methods. This difference in the accuracy and precision of parsimony and Bayesian approaches to topology estimation needs to be considered when selecting a method for phylogeny reconstruction.
Related collections
Most cited references6
- Record: found
- Abstract: found
- Article: not found
Divergence time estimation using fossils as terminal taxa and the origins of Lissamphibia.

- Record: found
- Abstract: found
- Article: found