- Record: found
- Abstract: found
- Article: found
Mass Spectrometric Identification of Urinary Biomarkers of Pulmonary Tuberculosis
Read this article at
Abstract
Background
Tuberculosis (TB) is the leading infectious cause of death worldwide. A major barrier to control of the pandemic is a lack of clinical biomarkers with the ability to distinguish active TB from healthy and sick controls and potential for development into point-of-care diagnostics.
Methods
We conducted a prospective case control study to identify candidate urine-based diagnostic biomarkers of active pulmonary TB (discovery cohort) and obtained a separate blinded “validation” cohort of confirmed cases of active pulmonary TB and controls with non-tuberculous pulmonary disease for validation. Clean-catch urine samples were collected and analyzed using high performance liquid chromatography-coupled time-of-flight mass spectrometry.
Results
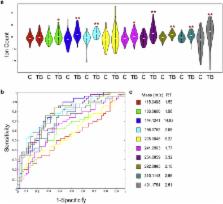
We discovered ten molecules from the discovery cohort with receiver-operator characteristic (ROC) area-under-the-curve (AUC) values >85%. These 10 molecules also significantly decreased after 60 days of treatment in a subset of 20 participants followed over time. Of these, a specific combination of diacetylspermine, neopterin, sialic acid, and N-acetylhexosamine exhibited ROC AUCs >80% in a blinded validation cohort of participants with active TB and non-tuberculous pulmonary disease.
Highlights
-
•
Urinary levels of small metabolites appear capable of distinguishing cases of active pulmonary tuberculosis from sick and healthy controls.
-
•
Levels of these biomarkers decrease after 60 days of treatment in a longitudinal cohort of 20 participants.
-
•
- Many of the identified biomarkers are known inflammatory intermediates that may reflect a specific immune response to tuberculosis.
Urine tests are commonly used to enable non-invasive, rapid and point-of-care diagnosis of various infectious diseases. We identified diacetylspermine, neopterin, sialic acid and N-acetylhexosamine as potential urine-based biomarkers for tuberculosis from two independent patient cohorts. These metabolites are known inflammatory intermediates and appear to decrease with anti-tuberculosis therapy in a subset of participants followed over 2 months. If validated, these metabolites have potential to both improve our understanding of the immune reaction to active tuberculosis and facilitate the development of a much-needed clinical biomarker for tuberculosis.
Related collections
Most cited references41
- Record: found
- Abstract: found
- Article: not found
Innovation: Metabolomics: the apogee of the omics trilogy.
- Record: found
- Abstract: found
- Article: not found
Xpert MTB/RIF assay for the diagnosis of extrapulmonary tuberculosis: a systematic review and meta-analysis.
- Record: found
- Abstract: found
- Article: not found