- Record: found
- Abstract: found
- Article: found
The Arabidopsis Golgi-localized GDP-L-fucose transporter is required for plant development

Read this article at
Abstract
Nucleotide sugar transport across Golgi membranes is essential for the luminal biosynthesis of glycan structures. Here we identify GDP-fucose transporter 1 (GFT1), an Arabidopsis nucleotide sugar transporter that translocates GDP- L-fucose into the Golgi lumen. Using proteo-liposome-based transport assays, we show that GFT preferentially transports GDP- L-fucose over other nucleotide sugars in vitro, while GFT1-silenced plants are almost devoid of L-fucose in cell wall-derived xyloglucan and rhamnogalacturonan II. Furthermore, these lines display reduced L-fucose content in N-glycan structures accompanied by severe developmental growth defects. We conclude that GFT1 is the major nucleotide sugar transporter for import of GDP- L-fucose into the Golgi and is required for proper plant growth and development.
Abstract
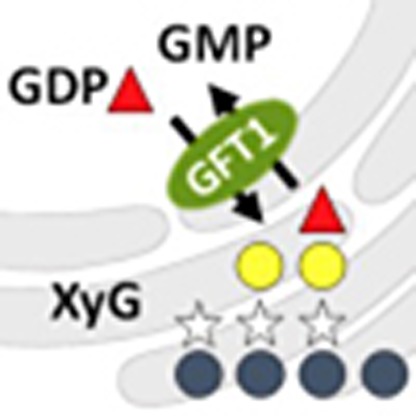 Nucleotide sugars are transported from the cytoplasm to the Golgi lumen where they
are incorporated into cell wall polysaccharides and used for glycosylation of proteins
and lipids. Here the authors identify GFT1, an
Arabidopsis Golgi-localized GDP-fucose transporter that is required for plant growth and development
Nucleotide sugars are transported from the cytoplasm to the Golgi lumen where they
are incorporated into cell wall polysaccharides and used for glycosylation of proteins
and lipids. Here the authors identify GFT1, an
Arabidopsis Golgi-localized GDP-fucose transporter that is required for plant growth and development
Related collections
Most cited references40
- Record: found
- Abstract: found
- Article: not found
NIH Image to ImageJ: 25 years of image analysis.
- Record: found
- Abstract: found
- Article: not found
Genome-wide insertional mutagenesis of Arabidopsis thaliana.
- Record: found
- Abstract: found
- Article: not found