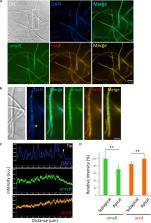
INTRODUCTION: The cytoplasm contains an unconventional class of organelles that concentrate specific factors and resources without a limiting membrane. These membraneless organelles include ribonucleoprotein (RNP) granules such as processing bodies (P-bodies, or PBs) and stress granules. PBs and stress granules are composed of nontranslating messenger RNAs (mRNAs) and associated proteins and are thought to provide discrete biochemical environments for regulating the translation and/or degradation of mRNA. In contrast to membrane-bound organelles, very little is known about what extrinsic and intrinsic factors regulate the fusion and fission of membrane-less organelles. Recently, an unexpected role for the endoplasmic reticulum (ER) has been observed in regulating the biogenesis of other membrane-bound organelles at contact sites where the two organelles are tethered and closely apposed. ER contact sites can allow the direct exchange of macromolecules and serve as a platform for the recruitment of machineries that regulate organelle biogenesis, division, and trafficking. Here, we found that ER contact sites can also regulate the biogenesis and fission of two types of membraneless organelles, PBs and stress granules. RATIONALE: To determine the extent to which PBs, a conserved cytoplasmic membraneless organelle, are tethered to the ER in animal cells, we used live-cell fluorescence microscopy to simultaneously track the spatiotemporal dynamics of the ER and PBs. To overcome the diffraction limits associated with light microscopy, we designed a reversible ER-PB contact assay using probes attached to the ER and PBs that emit a high-intensity fluorescence signal when the probes are close enough to dimerize. Because ER morphology and RNP granule biogenesis are tightly linked to mRNA translation, we systematically evaluated the relationships between ER morphology, RNP granule biogenesis, and mRNA translation by assessing endogenous PB numbers in response to altering ER shape and translational capacity and to the induction of cytosolic and ER stress. Because PBs and stress granules are dynamic organelles that undergo fission and fusion reactions akin to membrane-bound organelles, we used live-cell fluorescence microscopy to score the spatiotemporal relationship between the position of RNP granule division and contact sites with ER tubules. RESULTS: Using multiple measures, we found that a population of PBs were tethered to the ER in human cells. ER shape exerted profound effects on PB numbers and PB-ER contact. Conditions that promoted expansion of peripheral ER tubules and a reduction in peripheral ER cisternae increased PB numbers and ER-PB contact. Conversely, conditions that promoted an expansion of ER cisternae dramatically decreased PB numbers. The effect of ER shape on PB abundance was likely a reflection of the relative translational capacity of the ER domains. Owing to differences in ribosome density, smooth ER tubules are presumed to have a lower translational capacity than rough ER cisternae. Conditions that locally enhanced the translational capacity of the ER by increasing ER cisternae, such as ER stress, also reduced the number of PBs. Conversely, conditions that globally inhibited mRNA translation (NaAsO2 and puromycin) suppressed the effects of ER shape on PB abundance. Thus, ER contact sites affected the proliferation of PBs under basal and translationally repressed conditions. Furthermore, ER contact sites also affected the mysterious PB fission process. Live-cell imaging revealed that dynamic ER tubules define the position where PB and stress granule division occurs. These data mirror the spatiotemporal role of ER tubule contact domains that drive the constriction and division of membrane-bound organelles like endosomes and mitochondria. CONCLUSION: Here, we found that the ER contains contact site domains that are capable of tethering both membraneless and membrane-bound organelles. ER structure and translational capacity has effects on PB biogenesis. Furthermore, the fission of cytoplasmic RNP granules appears to represent an active process that can be driven by ER contact sites, analogous to the division of membrane-bound organelles. Endoplasmic reticulum tubules are a component of the ribonucleoprotein granule fission machinery. Membraneless RNP granules undergo fission and fusion similar to membrane-bound organelles. A cartoon (top) and the corresponding live-cell fluorescent images (bottom; at 0−, 5−, and 10-s time points, from left to right) of a PB (green) undergoing division at a position where an ER tubule is crossing (red). Tethered interactions between the endoplasmic reticulum (ER) and other membrane-bound organelles allow for efficient transfer of ions and/or macromolecules and provide a platform for organelle fission. Here, we describe an unconventional interface between membraneless ribonucleoprotein granules, such as processing bodies (P-bodies, or PBs) and stress granules, and the ER membrane. We found that PBs are tethered at molecular distances to the ER in human cells in a tunable fashion. ER-PB contact and PB biogenesis were modulated by altering PB composition, ER shape, or ER translational capacity. Furthermore, ER contact sites defined the position where PB and stress granule fission occurs. We thus suggest that the ER plays a fundamental role in regulating the assembly and disassembly of membraneless organelles.