- Record: found
- Abstract: found
- Article: found
Cytoplasmic TAF2–TAF8–TAF10 complex provides evidence for nuclear holo–TFIID assembly from preformed submodules

Read this article at
Abstract
General transcription factor TFIID is a cornerstone of RNA polymerase II transcription initiation in eukaryotic cells. How human TFIID—a megadalton-sized multiprotein complex composed of the TATA-binding protein (TBP) and 13 TBP-associated factors (TAFs)—assembles into a functional transcription factor is poorly understood. Here we describe a heterotrimeric TFIID subcomplex consisting of the TAF2, TAF8 and TAF10 proteins, which assembles in the cytoplasm. Using native mass spectrometry, we define the interactions between the TAFs and uncover a central role for TAF8 in nucleating the complex. X-ray crystallography reveals a non-canonical arrangement of the TAF8–TAF10 histone fold domains. TAF2 binds to multiple motifs within the TAF8 C-terminal region, and these interactions dictate TAF2 incorporation into a core–TFIID complex that exists in the nucleus. Our results provide evidence for a stepwise assembly pathway of nuclear holo–TFIID, regulated by nuclear import of preformed cytoplasmic submodules.
Abstract
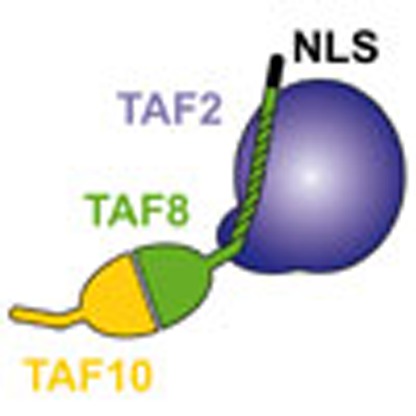 TFIID is an essential transcription factor complex that controls the expression of
most protein-coding genes in eukaryotes. Here the authors identify and characterize
a complex containing TAF2, TAF8 and TAF10, which assembles in the cytoplasm before
integration into the nuclear holo–TFIID complex.
TFIID is an essential transcription factor complex that controls the expression of
most protein-coding genes in eukaryotes. Here the authors identify and characterize
a complex containing TAF2, TAF8 and TAF10, which assembles in the cytoplasm before
integration into the nuclear holo–TFIID complex.
Related collections
Most cited references55
- Record: found
- Abstract: found
- Article: not found
ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins.
- Record: found
- Abstract: found
- Article: not found
A high-resolution map of active promoters in the human genome.
- Record: found
- Abstract: found
- Article: not found