- Record: found
- Abstract: found
- Article: not found
Genome-Wide Analysis of Cadmium-Induced, Germline Mutations in a Long-Term Daphnia pulex Mutation-Accumulation Experiment
Read this article at
Abstract
Background:
Germline mutations provide the raw material for all evolutionary processes and contribute to the occurrence of spontaneous human diseases and disorders. Yet despite the daily interaction of humans and other organisms with an increasing number of chemicals that are potentially mutagenic, precise measurements of chemically induced changes to the genome-wide rate and spectrum of germline mutation are lacking.
Objectives:
A large-scale Daphnia pulex mutation-accumulation experiment was propagated in the presence and absence of an environmentally relevant cadmium concentration to quantify the influence of cadmium on germline mutation rates and spectra.
Results:
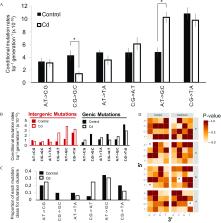
Cadmium exposure dramatically changed the genome-wide rates and regional spectra of germline mutations. In comparison with those in control conditions, Daphnia exposed to cadmium had a higher overall mutation rates and a lower overall mutation rate. Daphnia exposed to cadmium had a higher intergenic mutation rate and a lower exonic mutation rate. The higher intergenic mutation rate under cadmium exposure was the result of an elevated intergenic rate, whereas the lower exon mutation rate in cadmium was the result of a complete loss of exonic mutations—mutations that are known to be enriched at 5-hydroxymethylcytosine. We experimentally show that cadmium exposure significantly reduced 5-hydroxymethylcytosine levels.
Discussion:
These results provide evidence that cadmium changes regional mutation rates and can influence regional rates by interfering with an epigenetic process in the Daphnia pulex germline. We further suggest these observed cadmium-induced changes to the Daphnia germline mutation rate may be explained by cadmium’s inhibition of zinc-containing domains. The cadmium-induced changes to germline mutation rates and spectra we report provide a comprehensive view of the mutagenic perils of cadmium and give insight into its potential impact on human population health. https://doi.org/10.1289/EHP8932
Related collections
Most cited references56

- Record: found
- Abstract: found
- Article: found
Trimmomatic: a flexible trimmer for Illumina sequence data

- Record: found
- Abstract: found
- Article: found
The Sequence Alignment/Map format and SAMtools

- Record: found
- Abstract: found
- Article: found
