- Record: found
- Abstract: found
- Article: found
Bi-allelic loss-of-function variants in TMEM147 cause moderate to profound intellectual disability with facial dysmorphism and pseudo-Pelger-Huët anomaly
Read this article at
Summary
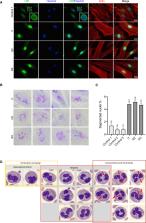
The transmembrane protein TMEM147 has a dual function: first at the nuclear envelope, where it anchors lamin B receptor (LBR) to the inner membrane, and second at the endoplasmic reticulum (ER), where it facilitates the translation of nascent polypeptides within the ribosome-bound TMCO1 translocon complex. Through international data sharing, we identified 23 individuals from 15 unrelated families with bi-allelic TMEM147 loss-of-function variants, including splice-site, nonsense, frameshift, and missense variants. These affected children displayed congruent clinical features including coarse facies, developmental delay, intellectual disability, and behavioral problems. In silico structural analyses predicted disruptive consequences of the identified amino acid substitutions on translocon complex assembly and/or function, and in vitro analyses documented accelerated protein degradation via the autophagy-lysosomal-mediated pathway. Furthermore, TMEM147-deficient cells showed CKAP4 (CLIMP-63) and RTN4 (NOGO) upregulation with a concomitant reorientation of the ER, which was also witnessed in primary fibroblast cell culture. LBR mislocalization and nuclear segmentation was observed in primary fibroblast cells. Abnormal nuclear segmentation and chromatin compaction were also observed in approximately 20% of neutrophils, indicating the presence of a pseudo-Pelger-Huët anomaly. Finally, co-expression analysis revealed significant correlation with neurodevelopmental genes in the brain, further supporting a role of TMEM147 in neurodevelopment. Our findings provide clinical, genetic, and functional evidence that bi-allelic loss-of-function variants in TMEM147 cause syndromic intellectual disability due to ER-translocon and nuclear organization dysfunction.
Graphical abstract
Abstract
TMEM147 plays an important role in protein localization and biogenesis. We discovered that individuals with bi-allelic TMEM147 loss-of-function variants show a neurodevelopmental disorder associated with facial dysmorphism and pseudo-Pelger-Huët anomaly. In primary cell lines, we observed nuclear envelope instability accompanied by lamin B receptor mislocalization and ER-translocon dysfunction.
Related collections
Most cited references54

- Record: found
- Abstract: found
- Article: found
CADD: predicting the deleteriousness of variants throughout the human genome
- Record: found
- Abstract: found
- Article: not found
REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants.
- Record: found
- Abstract: not found
- Article: not found
