- Record: found
- Abstract: found
- Article: found
Predicting provenance of forensic soil samples: Linking soil to ecological habitats by metabarcoding and supervised classification
Read this article at
Abstract
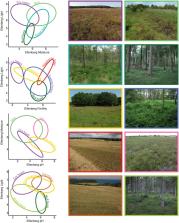
Environmental DNA (eDNA) is increasingly applied in ecological studies, including studies with the primary purpose of criminal investigation, in which eDNA from soil can be used to pair samples or reveal sample provenance. We collected soil eDNA samples as part of a large national biodiversity research project across 130 sites in Denmark. We investigated the potential for soil eDNA metabarcoding in predicting provenance in terms of environmental conditions, habitat type and geographic regions. We used linear regression for predicting environmental gradients of light, soil moisture, pH and nutrient status (represented by Ellenberg Indicator Values, EIVs) and Quadratic Discriminant Analysis (QDA) to predict habitat type and geographic region. eDNA data performed relatively well as a predictor of environmental gradients (R 2 > 0.81). Its ability to discriminate between habitat types was variable, with high accuracy for certain forest types and low accuracy for heathland, which was poorly predicted. Geographic region was also less accurately predicted by eDNA. We demonstrated the application of provenance prediction in forensic science by evaluating and discussing two mock crime scenes. Here, we listed the plant species from annotated sequences, which can further aid in identifying the likely habitat or, in case of rare species, a geographic region. Predictions of environmental gradients and habitat types together give an overall accurate description of a crime scene, but care should be taken when interpreting annotated sequences, e.g. due to erroneous assignments in GenBank. Our approach demonstrates that important habitat properties can be derived from soil eDNA, and exemplifies a range of potential applications of eDNA in forensic ecology.
Related collections
Most cited references25
- Record: found
- Abstract: not found
- Article: not found
Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data

- Record: found
- Abstract: found
- Article: found
Algorithm for post-clustering curation of DNA amplicon data yields reliable biodiversity estimates
- Record: found
- Abstract: found
- Article: not found