- Record: found
- Abstract: found
- Article: found
Phylogeny of saprobic microfungi from Southern Europe
Read this article at
Abstract
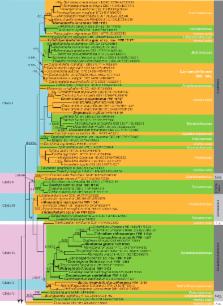
During a survey of saprophytic microfungi on decomposing woody, herbaceous debris and soil from different regions in Southern Europe, a wide range of interesting species of asexual ascomycetes were found. Phylogenetic analyses based on partial gene sequences of SSU, LSU and ITS proved that most of these fungi were related to Sordariomycetes and Dothideomycetes and to lesser extent to Leotiomycetes and Eurotiomycetes. Four new monotypic orders with their respective families are proposed here, i.e. Lauriomycetales, Lauriomycetaceae; Parasympodiellales, Parasympodiellaceae; Vermiculariopsiellales, Vermiculariopsiellaceae and Xenospadicoidales, Xenospadicoidaceae. One new order and three families are introduced here to accommodate orphan taxa, viz. Kirschsteiniotheliales, Castanediellaceae, Leptodontidiaceae and Pleomonodictydaceae. Furthermore, Bloxamiaceae is validated. Based on morphology and phylogenetic affinities Diplococcium singulare, Trichocladium opacum and Spadicoides atra are moved to the new genera Paradiplococcium, Pleotrichocladium and Xenospadicoides, respectively. Helicoon fuscosporum is accommodated in the genus Magnohelicospora. Other novel genera include Neoascotaiwania with the type species N. terrestris sp. nov., and N. limnetica comb. nov. previously accommodated in Ascotaiwania; Pleomonodictys with P. descalsii sp. nov. as type species, and P. capensis comb. nov. previously accommodated in Monodictys; Anapleurothecium typified by A. botulisporum sp. nov., a fungus morphologically similar to Pleurothecium but phylogenetically distant; Fuscosclera typified by F. lignicola sp. nov., a meristematic fungus related to Leotiomycetes; Pseudodiplococcium typified by P. ibericum sp. nov. to accommodate an isolate previously identified as Diplococcium pulneyense; Xyladictyochaeta typified with X. lusitanica sp. nov., a foliicolous fungus related to Xylariales and similar to Dictyochaeta, but distinguished by polyphialidic conidiogenous cells produced on setiform conidiophores. Other novel species proposed are Brachysporiella navarrica, Catenulostroma lignicola, Cirrenalia iberica, Conioscypha pleiomorpha, Leptodontidium aureum, Pirozynskiella laurisilvatica, Parasympodiella lauri and Zanclospora iberica. To fix the application of some fungal names, lectotypes and/or epitypes are designated for Magnohelicospora iberica, Sporidesmium trigonellum, Sporidesmium opacum, Sporidesmium asperum, Camposporium aquaticum and Psilonia atra.
Related collections
Most cited references136

- Record: found
- Abstract: found
- Article: found
One fungus, which genes? Development and assessment of universal primers for potential secondary fungal DNA barcodes

- Record: found
- Abstract: found
- Article: found
Towards an integrated phylogenetic classification of the Tremellomycetes

- Record: found
- Abstract: found
- Article: found