- Record: found
- Abstract: found
- Article: found
Towards unravelling Wolbachia global exchange: a contribution from the Bicyclus and Mylothris butterflies in the Afrotropics
Read this article at
Abstract
Background
Phylogenetically closely related strains of maternally inherited endosymbiotic bacteria are often found in phylogenetically divergent, and geographically distant insect host species. The interspecies transfer of the symbiont Wolbachia has been thought to have occurred repeatedly, facilitating its observed global pandemic. Few ecological interactions have been proposed as potential routes for the horizontal transfer of Wolbachia within natural insect communities. These routes are however likely to act only at the local scale, but how they may support the global distribution of some Wolbachia strains remains unclear.
Results
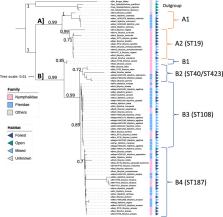
Here, we characterize the Wolbachia diversity in butterflies from the tropical forest regions of central Africa to discuss transfer at both local and global scales. We show that numerous species from both the Mylothris (family Pieridae) and Bicyclus (family Nymphalidae) butterfly genera are infected with similar Wolbachia strains, despite only minor interclade contacts across the life cycles of the species within their partially overlapping ecological niches. The phylogenetic distance and differences in resource use between these genera rule out the role of ancestry, hybridization, and shared host-plants in the interspecies transfer of the symbiont. Furthermore, we could not identify any shared ecological factors to explain the presence of the strains in other arthropod species from other habitats, or even ecoregions.
Related collections
Most cited references59
- Record: found
- Abstract: found
- Article: found
RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies

- Record: found
- Abstract: found
- Article: found
Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data

- Record: found
- Abstract: found
- Article: found