- Record: found
- Abstract: found
- Article: found
Multiple Plant Surface Signals are Sensed by Different Mechanisms in the Rice Blast Fungus for Appressorium Formation
Read this article at
Abstract
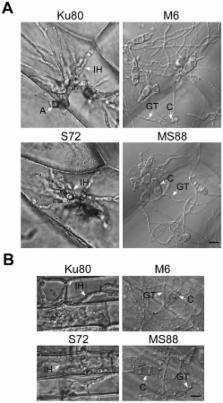
Surface recognition and penetration are among the most critical plant infection processes in foliar pathogens. In Magnaporthe oryzae, the Pmk1 MAP kinase regulates appressorium formation and penetration. Its orthologs also are known to be required for various plant infection processes in other phytopathogenic fungi. Although a number of upstream components of this important pathway have been characterized, the upstream sensors for surface signals have not been well characterized. Pmk1 is orthologous to Kss1 in yeast that functions downstream from Msb2 and Sho1 for filamentous growth. Because of the conserved nature of the Pmk1 and Kss1 pathways and reduced expression of MoMSB2 in the pmk1 mutant, in this study we functionally characterized the MoMSB2 and MoSHO1 genes. Whereas the Momsb2 mutant was significantly reduced in appressorium formation and virulence, the Mosho1 mutant was only slightly reduced. The Mosho1 Momsb2 double mutant rarely formed appressoria on artificial hydrophobic surfaces, had a reduced Pmk1 phosphorylation level, and was nonresponsive to cutin monomers. However, it still formed appressoria and caused rare, restricted lesions on rice leaves. On artificial hydrophilic surfaces, leaf surface waxes and primary alcohols-but not paraffin waxes and alkanes- stimulated appressorium formation in the Mosho1 Momsb2 mutant, but more efficiently in the Momsb2 mutant. Furthermore, expression of a dominant active MST7 allele partially suppressed the defects of the Momsb2 mutant. These results indicate that, besides surface hydrophobicity and cutin monomers, primary alcohols, a major component of epicuticular leaf waxes in grasses, are recognized by M. oryzae as signals for appressorium formation. Our data also suggest that MoMsb2 and MoSho1 may have overlapping functions in recognizing various surface signals for Pmk1 activation and appressorium formation. While MoMsb2 is critical for sensing surface hydrophobicity and cutin monomers, MoSho1 may play a more important role in recognizing rice leaf waxes.
Author Summary
The rice blast fungus is a major pathogen of rice and a model for studying fungal-plant interactions. Like many other fungal pathogens, it can recognize physical and chemical signals present on the rice leaf surface and form a highly specialized infection structure known as appressorium. A well conserved signal transduction pathway involving the protein kinase gene PMK1 is known to regulate appressorium formation and plant penetration in this pathogen. However, it is not clear about the sensor genes that are involved in recognizing various plant surface signals. In this study we functionally characterize two putative sensor genes called MoMSB2 and MoSHO1. Genetic and biochemical analyses indicated that these two genes have overlapping functions in recognizing different physical and chemical signals present on the rice leaf surface for the activation of the Pmk1 pathway and appressorium formation. We found that primary alcohols, a major component of leaf waxes in grasses, can be recognized by the rice blast fungus as chemical cues. While MoMSB2 is critical for sensing hydrophobicity and precursors of cutin molecules of rice leaves, MoSHO1 appears to be more important than MoMSB2 for recognizing wax components.
Related collections
Most cited references41
- Record: found
- Abstract: found
- Article: not found
Structure and function of the cell surface (tethered) mucins.
- Record: found
- Abstract: found
- Article: not found
Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea.
- Record: found
- Abstract: found
- Article: not found