- Record: found
- Abstract: found
- Article: found
Multi-layered prevention and treatment of chronic inflammation, organ fibrosis and cancer associated with canonical WNT/β-catenin signaling activation (Review)

Read this article at
Abstract
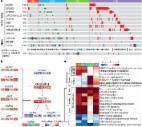
β-catenin/CTNNB1 is an intracellular scaffold protein that interacts with adhesion molecules (E-cadherin/CDH1, N-cadherin/CDH2, VE-cadherin/CDH5 and α-catenins), transmembrane-type mucins (MUC1/CD227 and MUC16/CA125), signaling regulators (APC, AXIN1, AXIN2 and NHERF1/EBP50) and epigenetic or transcriptional regulators (BCL9, BCL9L, CREBBP/CBP, EP300/p300, FOXM1, MED12, SMARCA4/BRG1 and TCF/LEF). Gain-of-function CTTNB1 mutations are detected in bladder cancer, colorectal cancer, gastric cancer, liver cancer, lung cancer, pancreatic cancer, prostate cancer and uterine cancer, whereas loss-of-function CTNNB1 mutations are also detected in human cancer. ABCB1, ALDH1A1, ASCL2, ATF3, AXIN2, BAMBI, CCND1, CD44, CLDN1, CTLA4, DKK1, EDN1, EOMES, FGF18, FGF20, FZD7, IL10, JAG1, LEF1, LGR5, MITF, MSX1, MYC, NEUROD1, NKD1, NODAL, NOTCH2, NOTUM, NRCAM, OPN, PAX3, PPARD, PTGS2, RNF43, SNAI1, SP5, TCF7, TERT, TNFRSF19, VEGFA and ZNRF3 are representative β-catenin target genes. β-catenin signaling is involved in myofibroblast activation and subsequent pulmonary fibrosis, in addition to other types of fibrosis. β-catenin and NF-κB signaling activation are involved in field cancerization in the stomach associated with Helicobacter pylori ( H. pylori) infection and in the liver associated with hepatitis C virus (HCV) infection and other etiologies. β-catenin-targeted therapeutics are functionally classified into β-catenin inhibitors targeting upstream regulators (AZ1366, ETC-159, G007-LK, GNF6231, ipafricept, NVP-TNKS656, rosmantuzumab, vantictumab, WNT-C59, WNT974 and XAV939), β-catenin inhibitors targeting protein-protein interactions (CGP049090, CWP232228, E7386, ICG-001, LF3 and PRI-724), β-catenin inhibitors targeting epigenetic regulators (PKF118-310), β-catenin inhibitors targeting mediator complexes (CCT251545 and cortistatin A) and β-catenin inhibitors targeting transmembrane-type transcriptional outputs, including CD44v6, FZD7 and LGR5. Eradicating H. pylori and HCV is the optimal approach for the first-line prevention of gastric cancer and hepatocellular carcinoma (HCC), respectively. However, β-catenin inhibitors may be applicable for the prevention of organ fibrosis, second-line HCC prevention and treating β-catenin-driven cancer. The multi-layered prevention and treatment strategy of β-catenin-related human diseases is necessary for the practice of personalized medicine and implementation of precision medicine.
Related collections
Most cited references168
- Record: found
- Abstract: found
- Article: found
Comprehensive molecular characterization of gastric adenocarcinoma
- Record: found
- Abstract: found
- Article: not found
Liver fibrosis.
- Record: found
- Abstract: found
- Article: not found