- Record: found
- Abstract: found
- Article: found
Deep learning for chest radiograph diagnosis: A retrospective comparison of the CheXNeXt algorithm to practicing radiologists
Read this article at
Abstract
Background
Chest radiograph interpretation is critical for the detection of thoracic diseases, including tuberculosis and lung cancer, which affect millions of people worldwide each year. This time-consuming task typically requires expert radiologists to read the images, leading to fatigue-based diagnostic error and lack of diagnostic expertise in areas of the world where radiologists are not available. Recently, deep learning approaches have been able to achieve expert-level performance in medical image interpretation tasks, powered by large network architectures and fueled by the emergence of large labeled datasets. The purpose of this study is to investigate the performance of a deep learning algorithm on the detection of pathologies in chest radiographs compared with practicing radiologists.
Methods and findings
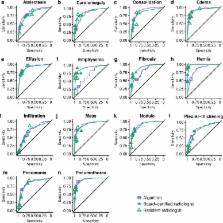
We developed CheXNeXt, a convolutional neural network to concurrently detect the presence of 14 different pathologies, including pneumonia, pleural effusion, pulmonary masses, and nodules in frontal-view chest radiographs. CheXNeXt was trained and internally validated on the ChestX-ray8 dataset, with a held-out validation set consisting of 420 images, sampled to contain at least 50 cases of each of the original pathology labels. On this validation set, the majority vote of a panel of 3 board-certified cardiothoracic specialist radiologists served as reference standard. We compared CheXNeXt’s discriminative performance on the validation set to the performance of 9 radiologists using the area under the receiver operating characteristic curve (AUC). The radiologists included 6 board-certified radiologists (average experience 12 years, range 4–28 years) and 3 senior radiology residents, from 3 academic institutions. We found that CheXNeXt achieved radiologist-level performance on 11 pathologies and did not achieve radiologist-level performance on 3 pathologies. The radiologists achieved statistically significantly higher AUC performance on cardiomegaly, emphysema, and hiatal hernia, with AUCs of 0.888 (95% confidence interval [CI] 0.863–0.910), 0.911 (95% CI 0.866–0.947), and 0.985 (95% CI 0.974–0.991), respectively, whereas CheXNeXt’s AUCs were 0.831 (95% CI 0.790–0.870), 0.704 (95% CI 0.567–0.833), and 0.851 (95% CI 0.785–0.909), respectively. CheXNeXt performed better than radiologists in detecting atelectasis, with an AUC of 0.862 (95% CI 0.825–0.895), statistically significantly higher than radiologists' AUC of 0.808 (95% CI 0.777–0.838); there were no statistically significant differences in AUCs for the other 10 pathologies. The average time to interpret the 420 images in the validation set was substantially longer for the radiologists (240 minutes) than for CheXNeXt (1.5 minutes). The main limitations of our study are that neither CheXNeXt nor the radiologists were permitted to use patient history or review prior examinations and that evaluation was limited to a dataset from a single institution.
Conclusions
In this study, we developed and validated a deep learning algorithm that classified clinically important abnormalities in chest radiographs at a performance level comparable to practicing radiologists. Once tested prospectively in clinical settings, the algorithm could have the potential to expand patient access to chest radiograph diagnostics.
Abstract
In their study, Pranav Rajpurkar and colleagues test a deep learning algorithm that classifies clinically important abnormalities in chest radiographs.
Author summary
Why was this study done?
-
Chest radiographs are the most common medical imaging test in the world and critical for diagnosing common thoracic diseases.
-
Radiograph interpretation is a time-consuming task, and there is shortage of qualified trained radiologists in many healthcare systems.
-
Deep learning algorithms that have been developed to provide diagnostic chest radiograph interpretation have not been compared to expert human radiologist performance.
What did the researchers do and find?
-
We developed a deep learning algorithm to concurrently detect 14 clinically important pathologies in chest radiographs.
-
The algorithm can also localize parts of the image most indicative of each pathology.
-
We evaluated the algorithm against 9 practicing radiologists on a validation set of 420 images for which the majority vote of 3 cardiothoracic specialty radiologists served as ground truth.
-
The algorithm achieved performance equivalent to the practicing radiologists on 10 pathologies, better on 1 pathology, and worse on 3 pathologies.
-
Radiologists labeled the 420 images in 240 minutes on average, and the algorithm labeled them in 1.5 minutes.
What do these findings mean?
-
Deep learning algorithms can diagnose certain pathologies in chest radiographs at a level comparable to practicing radiologists on a single institution dataset.
-
After clinical validation, algorithms such as the one presented in this work could be used to increase access to rapid, high-quality chest radiograph interpretation.
Related collections
Most cited references32
- Record: found
- Abstract: not found
- Conference Proceedings: not found
Learning Deep Features for Discriminative Localization
- Record: found
- Abstract: found
- Article: not found
Training and Validating a Deep Convolutional Neural Network for Computer-Aided Detection and Classification of Abnormalities on Frontal Chest Radiographs.
- Record: found
- Abstract: found
- Article: not found