- Record: found
- Abstract: found
- Article: found
RNA-on-X 1 and 2 in Drosophila melanogaster fulfill separate functions in dosage compensation
Read this article at
Abstract
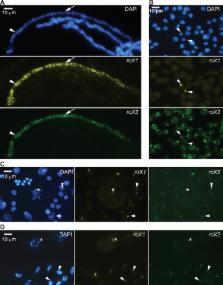
In Drosophila melanogaster, the male-specific lethal (MSL) complex plays a key role in dosage compensation by stimulating expression of male X-chromosome genes. It consists of MSL proteins and two long noncoding RNAs, roX1 and roX2, that are required for spreading of the complex on the chromosome and are redundant in the sense that loss of either does not affect male viability. However, despite rapid evolution, both roX species are present in diverse Drosophilidae species, raising doubts about their full functional redundancy. Thus, we have investigated consequences of deleting roX1 and/or roX2 to probe their specific roles and redundancies in D. melanogaster. We have created a new mutant allele of roX2 and show that roX1 and roX2 have partly separable functions in dosage compensation. In larvae, roX1 is the most abundant variant and the only variant present in the MSL complex when the complex is transmitted (physically associated with the X-chromosome) in mitosis. Loss of roX1 results in reduced expression of the genes on the X-chromosome, while loss of roX2 leads to MSL-independent upregulation of genes with male-biased testis-specific transcription. In roX1 roX2 mutant, gene expression is strongly reduced in a manner that is not related to proximity to high-affinity sites. Our results suggest that high tolerance of mis-expression of the X-chromosome has evolved. We propose that this may be a common property of sex-chromosomes, that dosage compensation is a stochastic process and its precision for each individual gene is regulated by the density of high-affinity sites in the locus.
Author summary
In humans and fruit flies, females and males have different sets of sex chromosomes. This causes gene dosage differences that must be compensated for by adjusting the expression of most genes located on the X-chromosome. Long non-coding RNAs are central in this compensation and in fruit flies this is mediated by two non-coding RNAs, roX1 and roX2 which together with five proteins form the male-specific lethal complex. The complex recognizes and upregulates gene transcription on the male X-chromosome. While non-coding RNAs are are engaged in numerous biological processes and critical for compensation their precise functions remain elusive. To understand the function of long non-coding RNAs we analysed the expression of all genes in roX1, roX2 and roX1 roX2 mutants to explore the roles of long non-coding RNAs. These mutants have different impacts on the genome-wide expression. Our results also suggest that the X-chromosome is highly tolerant to mis-expression and we speculate that this tolerance evolved in parallel with compensation mechanisms and may be a common property of sex-chromosomes. We propose that dosage compensation is a stochastic process that depends on the distribution of specific binding sites which will be selected for and optimized depending on the genes’ individual expression levels.
Related collections
Most cited references77
- Record: found
- Abstract: found
- Article: not found
Highly improved gene targeting by germline-specific Cas9 expression in Drosophila.
- Record: found
- Abstract: found
- Article: not found
Evolution of genetic redundancy.

- Record: found
- Abstract: found
- Article: found