- Record: found
- Abstract: found
- Article: found
Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice
Read this article at
Abstract
Structure of a bacterial adhesin reveals its role in forming a mixed-species symbiotic community with diatoms on sea ice.
Abstract
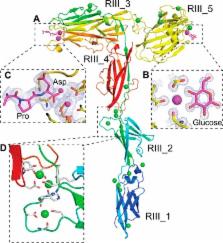
Bacterial adhesins are modular cell-surface proteins that mediate adherence to other cells, surfaces, and ligands. The Antarctic bacterium Marinomonas primoryensis uses a 1.5-MDa adhesin comprising over 130 domains to position it on ice at the top of the water column for better access to oxygen and nutrients. We have reconstructed this 0.6-μm-long adhesin using a “dissect and build” structural biology approach and have established complementary roles for its five distinct regions. Domains in region I (RI) tether the adhesin to the type I secretion machinery in the periplasm of the bacterium and pass it through the outer membrane. RII comprises ~120 identical immunoglobulin-like β-sandwich domains that rigidify on binding Ca 2+ to project the adhesion regions RIII and RIV into the medium. RIII contains ligand-binding domains that join diatoms and bacteria together in a mixed-species community on the underside of sea ice where incident light is maximal. RIV is the ice-binding domain, and the terminal RV domain contains several “repeats-in-toxin” motifs and a noncleavable signal sequence that target proteins for export via the type I secretion system. Similar structural architecture is present in the adhesins of many pathogenic bacteria and provides a guide to finding and blocking binding domains to weaken infectivity.
Related collections
Most cited references40
- Record: found
- Abstract: found
- Article: not found
Scaling and assessment of data quality
- Record: found
- Abstract: found
- Article: not found
REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use.
- Record: found
- Abstract: found
- Article: not found