- Record: found
- Abstract: found
- Article: not found
IFITMs Restrict the Replication of Multiple Pathogenic Viruses
Abstract
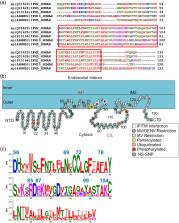
The interferon-inducible transmembrane protein (IFITM) family inhibits a growing number of pathogenic viruses, among them influenza A virus, dengue virus, hepatitis C virus, and Ebola virus. This review covers recent developments in our understanding of the IFITM's molecular determinants, potential mechanisms of action, and impact on pathogenesis.
Graphical abstract
Highlights
-
•
IFITMs are host intramembrane proteins that block viral fusion resulting in virus degradation.
-
•
IFITMs inhibit a growing number of pathogenic viruses, including influenza A virus, dengue virus, and Ebola virus.
-
•
Mice null for Ifitm3 exhibit increased disease severity when infected with the influenza A virus.
-
•
Patients expressing a variant allele of IFITM3 are more likely to develop severe influenza infection.
-
•
Recent work suggests that IFITMs may toughen the host membrane, thus preventing viral membrane fusion.
Related collections
Most cited references108
- Record: found
- Abstract: found
- Article: not found
Systematic and quantitative assessment of the ubiquitin-modified proteome.
- Record: found
- Abstract: found
- Article: not found
Signals for sorting of transmembrane proteins to endosomes and lysosomes.
- Record: found
- Abstract: found
- Article: not found

