- Record: found
- Abstract: found
- Article: found
A Multiple Peptides Vaccine against COVID-19 Designed from the Nucleocapsid phosphoprotein (N) and Spike Glycoprotein (S) via the Immunoinformatics Approach
Read this article at
Abstract
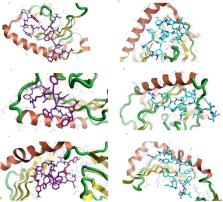
Due to the current Coronavirus (COVID-19) pandemic, the rapid discovery of a safe and effective vaccine is an essential issue. Consequently, this study aims to predict a potential COVID-19 peptide-based vaccine utilizing the Nucleocapsid phosphoprotein (N) and Spike Glycoprotein (S) via the Immunoinformatics approach. To achieve this goal, several Immune Epitope Database (IEDB) tools, molecular docking, and safety prediction servers were used. According to the results, The Spike peptide SQCVNLTTRTQLPPAYTNSFTRGVY is predicted to have the highest binding affinity to the B-Cells. The Spike peptide FTISVTTEI has the highest binding affinity to the Major Histocompatibility Complex class 1 (MHC I) Human Leukocyte Allele HLA-B*1503 (according to the MDockPeP and HPEPDOCK servers, docking scores were -153.9 and -229.356, respectively). The Nucleocapsid peptides KTFPPTEPK and RWYFYYLGTGPEAGL have the highest binding affinity to the MHC I HLA-A0202 allele and the three the Major Histocompatibility Complex class 2 (MHC II) Human Leukocyte Allele HLA-DPA1*01:03/DPB1*02:01, HLA-DQA1*01:02/DQB1-*06:02, HLA-DRB1, respectively. Docking scores of peptide KTFPPTEPK were -153.9 and -220.876. In contrast, docking scores of peptide RWYFYYLGTGPEAGL were ranged from 218-318. Furthermore, those peptides were predicted as non-toxic and non-allergen. Therefore, the combination of those peptides is predicted to stimulate better immunological responses with respectable safety.
Related collections
Most cited references47
- Record: found
- Abstract: found
- Article: not found
UCSF Chimera--a visualization system for exploratory research and analysis.
- Record: found
- Abstract: found
- Article: not found
The Protein Data Bank.
- Record: found
- Abstract: found
- Article: not found
