- Record: found
- Abstract: found
- Article: found
clusterProfiler 4.0: A universal enrichment tool for interpreting omics data

Read this article at
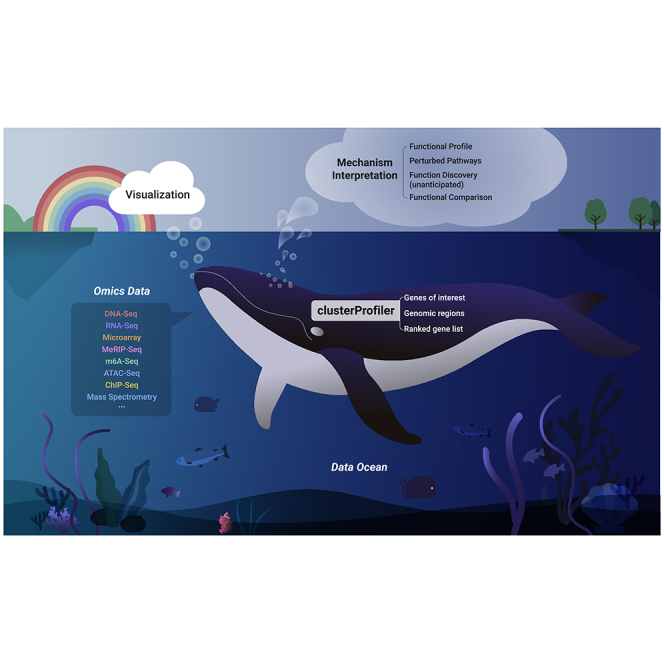
Summary
Functional enrichment analysis is pivotal for interpreting high-throughput omics data in life science. It is crucial for this type of tool to use the latest annotation databases for as many organisms as possible. To meet these requirements, we present here an updated version of our popular Bioconductor package, clusterProfiler 4.0. This package has been enhanced considerably compared with its original version published 9 years ago. The new version provides a universal interface for functional enrichment analysis in thousands of organisms based on internally supported ontologies and pathways as well as annotation data provided by users or derived from online databases. It also extends the dplyr and ggplot2 packages to offer tidy interfaces for data operation and visualization. Other new features include gene set enrichment analysis and comparison of enrichment results from multiple gene lists. We anticipate that clusterProfiler 4.0 will be applied to a wide range of scenarios across diverse organisms.
Graphical abstract
Public summary
-
•
clusterProfiler supports exploring functional characteristics of both coding and non-coding genomics data for thousands of species with up-to-date gene annotation
-
•
It provides a universal interface for gene functional annotation from a variety of sources and thus can be applied in diverse scenarios
-
•
It provides a tidy interface to access, manipulate, and visualize enrichment results to help users achieve efficient data interpretation
-
•
Datasets obtained from multiple treatments and time points can be analyzed and compared in a single run, easily revealing functional consensus and differences among distinct conditions
Related collections
Most cited references43
- Record: found
- Abstract: found
- Article: not found
Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles
- Record: found
- Abstract: found
- Article: not found
clusterProfiler: an R package for comparing biological themes among gene clusters.

- Record: found
- Abstract: found
- Article: found