- Record: found
- Abstract: found
- Article: not found
HDAC inhibitors attenuate the development of hypersensitivity in models of neuropathic pain
Read this article at
Summary
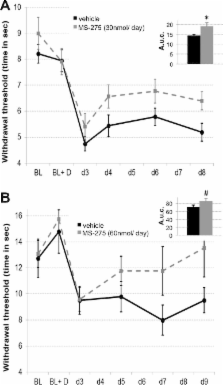
Intrathecal delivery of histone deacetylase inhibitors ameliorates hypersensitivity in models of neuropathic pain. This effect may be mediated at the level of the spinal cord through inhibition of HDAC1 function.
Abstract
Histone deacetylase inhibitors (HDACIs) interfere with the epigenetic process of histone acetylation and are known to have analgesic properties in models of chronic inflammatory pain. The aim of this study was to determine whether these compounds could also affect neuropathic pain. Different class I HDACIs were delivered intrathecally into rat spinal cord in models of traumatic nerve injury and antiretroviral drug–induced peripheral neuropathy (stavudine, d4T). Mechanical and thermal hypersensitivity was attenuated by 40% to 50% as a result of HDACI treatment, but only if started before any insult. The drugs globally increased histone acetylation in the spinal cord, but appeared to have no measurable effects in relevant dorsal root ganglia in this treatment paradigm, suggesting that any potential mechanism should be sought in the central nervous system. Microarray analysis of dorsal cord RNA revealed the signature of the specific compound used (MS-275) and suggested that its main effect was mediated through HDAC1. Taken together, these data support a role for histone acetylation in the emergence of neuropathic pain.
Related collections
Most cited references47
- Record: found
- Abstract: found
- Article: not found
Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt.
- Record: found
- Abstract: found
- Article: not found
A new and sensitive method for measuring thermal nociception in cutaneous hyperalgesia.
- Record: found
- Abstract: found
- Article: not found
