- Record: found
- Abstract: found
- Article: found
Contribution of epigenetic landscapes and transcription factors to X-chromosome reactivation in the inner cell mass
Read this article at
Abstract
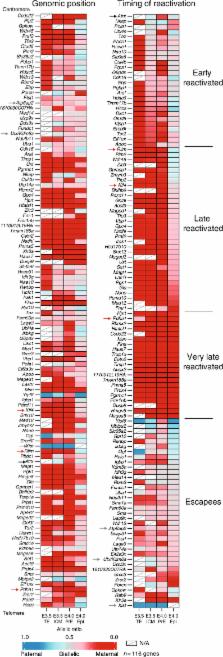
X-chromosome inactivation is established during early development. In mice, transcriptional repression of the paternal X-chromosome (Xp) and enrichment in epigenetic marks such as H3K27me3 is achieved by the early blastocyst stage. X-chromosome inactivation is then reversed in the inner cell mass. The mechanisms underlying Xp reactivation remain enigmatic. Using in vivo single-cell approaches (allele-specific RNAseq, nascent RNA-fluorescent in situ hybridization and immunofluorescence), we show here that different genes are reactivated at different stages, with more slowly reactivated genes tending to be enriched in H3meK27. We further show that in UTX H3K27 histone demethylase mutant embryos, these genes are even more slowly reactivated, suggesting that these genes carry an epigenetic memory that may be actively lost. On the other hand, expression of rapidly reactivated genes may be driven by transcription factors. Thus, some X-linked genes have minimal epigenetic memory in the inner cell mass, whereas others may require active erasure of chromatin marks.
Abstract
X-chromosome inactivation is reversed in the mouse inner cell mass (ICM) through a mechanism that is not fully understood. Here, the authors investigate this process and characterize the contributions of the epigenetic landscape and transcription factors in X-linked gene reactivation dynamics.
Related collections
Most cited references41
- Record: found
- Abstract: found
- Article: not found
Gene Ontology: tool for the unification of biology
- Record: found
- Abstract: not found
- Article: not found
Gene action in the X-chromosome of the mouse (Mus musculus L.).
- Record: found
- Abstract: found
- Article: not found