- Record: found
- Abstract: found
- Article: found
Population variability of rhesus macaque ( Macaca mulatta) NAT1 gene for arylamine N-acetyltransferase 1: Functional effects and comparison with human
Read this article at
Abstract
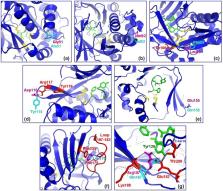
Human NAT1 gene for N-acetyltransferase 1 modulates xenobiotic metabolism of arylamine drugs and mutagens. Beyond pharmacogenetics, NAT1 is also relevant to breast cancer. The population history of human NAT1 suggests evolution through purifying selection, but it is unclear whether this pattern is evident in other primate lineages where population studies are scarce. We report NAT1 polymorphism in 25 rhesus macaques ( Macaca mulatta) and describe the haplotypic and functional characteristics of 12 variants. Seven non-synonymous single nucleotide variations (SNVs) were identified and experimentally demonstrated to compromise enzyme function, mainly through destabilization of NAT1 protein and consequent activity loss. One non-synonymous SNV (c.560G > A, p.Arg187Gln) has also been characterized for human NAT1 with similar effects. Population haplotypic and functional variability of rhesus NAT1 was considerably higher than previously reported for its human orthologue, suggesting different environmental pressures in the two lineages. Known functional elements downstream of human NAT1 were also differentiated in rhesus macaque and other primates. Xenobiotic metabolizing enzymes play roles beyond mere protection from exogenous chemicals. Therefore, any link to disease, particularly carcinogenesis, may be via modulation of xenobiotic mutagenicity or more subtle interference with cell physiology. Comparative analyses add the evolutionary dimension to such investigations, assessing functional conservation/diversification among primates.
Related collections
Most cited references105
- Record: found
- Abstract: found
- Article: not found
A database of vertebrate longevity records and their relation to other life-history traits.

- Record: found
- Abstract: found
- Article: found