- Record: found
- Abstract: found
- Article: found
Microbiota alteration is associated with the development of stress-induced despair behavior
Read this article at
Abstract
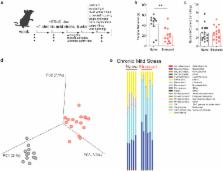
Depressive disorders often run in families, which, in addition to the genetic component, may point to the microbiome as a causative agent. Here, we employed a combination of behavioral, molecular and computational techniques to test the role of the microbiota in mediating despair behavior. In chronically stressed mice displaying despair behavior, we found that the microbiota composition and the metabolic signature dramatically change. Specifically, we observed reduced Lactobacillus and increased circulating kynurenine levels as the most prominent changes in stressed mice. Restoring intestinal Lactobacillus levels was sufficient to improve the metabolic alterations and behavioral abnormalities. Mechanistically, we identified that Lactobacillus-derived reactive oxygen species may suppress host kynurenine metabolism, by inhibiting the expression of the metabolizing enzyme, IDO1, in the intestine. Moreover, maintaining elevated kynurenine levels during Lactobacillus supplementation diminished the treatment benefits. Collectively, our data provide a mechanistic scenario for how a microbiota player ( Lactobacillus) may contribute to regulating metabolism and resilience during stress.
Related collections
Most cited references46
- Record: found
- Abstract: found
- Article: not found
FLASH: fast length adjustment of short reads to improve genome assemblies.
- Record: found
- Abstract: found
- Article: not found
Housekeeping genes as internal standards: use and limits.
- Record: found
- Abstract: found
- Article: not found