- Record: found
- Abstract: found
- Article: found
Characterisation of the active/de-active transition of mitochondrial complex I ☆

Read this article at
Abstract
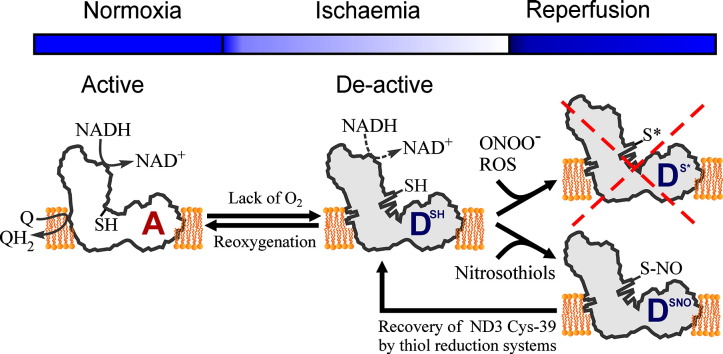
Oxidation of NADH in the mitochondrial matrix of aerobic cells is catalysed by mitochondrial complex I. The regulation of this mitochondrial enzyme is not completely understood. An interesting characteristic of complex I from some organisms is the ability to adopt two distinct states: the so-called catalytically active (A) and the de-active, dormant state (D). The A-form in situ can undergo de-activation when the activity of the respiratory chain is limited (i.e. in the absence of oxygen).
The mechanisms and driving force behind the A/D transition of the enzyme are currently unknown, but several subunits are most likely involved in the conformational rearrangements: the accessory subunit 39 kDa (NDUFA9) and the mitochondrially encoded subunits, ND3 and ND1. These three subunits are located in the region of the quinone binding site.
The A/D transition could represent an intrinsic mechanism which provides a fast response of the mitochondrial respiratory chain to oxygen deprivation. The physiological role of the accumulation of the D-form in anoxia is most probably to protect mitochondria from ROS generation due to the rapid burst of respiration following reoxygenation. The de-activation rate varies in different tissues and can be modulated by the temperature, the presence of free fatty acids and divalent cations, the NAD +/NADH ratio in the matrix, the presence of nitric oxide and oxygen availability.
Cysteine-39 of the ND3 subunit, exposed in the D-form, is susceptible to covalent modification by nitrosothiols, ROS and RNS. The D-form in situ could react with natural effectors in mitochondria or with pharmacological agents. Therefore the modulation of the re-activation rate of complex I could be a way to ameliorate the ischaemia/reperfusion damage. This article is part of a Special Issue entitled: 18th European Bioenergetic Conference. Guest Editors: Manuela Pereira and Miguel Teixeira.
Graphical abstract
Highlights
Related collections
Most cited references137
- Record: found
- Abstract: found
- Article: not found
Supercomplexes in the respiratory chains of yeast and mammalian mitochondria.
- Record: found
- Abstract: found
- Article: not found
Supercomplex assembly determines electron flux in the mitochondrial electron transport chain.
- Record: found
- Abstract: found
- Article: not found