- Record: found
- Abstract: found
- Article: found
Selection and Characterization of a Novel DNA Aptamer, Apt-07S Specific to Hepatocellular Carcinoma Cells
Abstract
Background
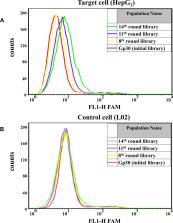
The efficacy of traditional therapeutic methods for liver cancer is unsatisfying because of the poor targeting, and inefficient drug delivery system. A recent study has proven that aptamers, developed through cell-SELEX, could specifically recognize cancer cells and show great potential in the development of a delivery system for anticancer drugs.
Purpose
To develop a hepatocellular carcinoma specific aptamer using two kinds of hepatocellular carcinoma cell lines, HepG 2 and SMMC-7721, as double targets and a normal hepatocyte, L02, as a negative control cell.
Methods
Hepatocellular carcinoma specific aptamer was developed via cell-SELEX. The enrichment of the library was monitored by flow cytometric analysis. The specificity, affinity, and distribution of the candidate aptamer were explored. Further study was carried to assess its potential in drug delivery.
Results
The library was enriched after 14 rounds of screening. Candidate aptamer Apt-07S can recognize four kinds of hepatocellular carcinoma cells and show little cell-binding ability to normal cells and four cell lines of different cancer types, revealing a high specificity of Apt-07S. Confocal imaging showed that Apt-07S distributed both on the surface and in the cytoplasm of the two target cells. Moreover, an anti-sense nucleotide to gene Plk1 (ASO-Plk1) was connected at the 3ʹ end of Apt-07S to form an integrated molecule (Apt-07S-ASO-Plk1); the functional analysis indicated that the structure of Apt-07S may help ASO-Plk1 enter the cancer cells.
Most cited references32

- Record: found
- Abstract: found
- Article: found
Recent Advances in Aptamer Discovery and Applications
- Record: found
- Abstract: found
- Article: not found
CD133 positive hepatocellular carcinoma cells possess high capacity for tumorigenicity.
- Record: found
- Abstract: not found
- Article: not found