- Record: found
- Abstract: found
- Article: found
Genomic, Transcriptomic and Metabolomic Studies of Two Well-Characterized, Laboratory-Derived Vancomycin-Intermediate Staphylococcus aureus Strains Derived from the Same Parent Strain
Read this article at
Abstract
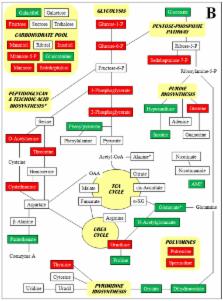
Complete genome comparisons, transcriptomic and metabolomic studies were performed on two laboratory-selected, well-characterized vancomycin-intermediate Staphylococcus aureus (VISA) derived from the same parent MRSA that have changes in cell wall composition and decreased autolysis. A variety of mutations were found in the VISA, with more in strain 13136p −m +V20 (vancomycin MIC = 16 µg/mL) than strain 13136p −m +V5 (MIC = 8 µg/mL). Most of the mutations have not previously been associated with the VISA phenotype; some were associated with cell wall metabolism and many with stress responses, notably relating to DNA damage. The genomes and transcriptomes of the two VISA support the importance of gene expression regulation to the VISA phenotype. Similarities in overall transcriptomic and metabolomic data indicated that the VISA physiologic state includes elements of the stringent response, such as downregulation of protein and nucleotide synthesis, the pentose phosphate pathway and nutrient transport systems. Gene expression for secreted virulence determinants was generally downregulated, but was more variable for surface-associated virulence determinants, although capsule formation was clearly inhibited. The importance of activated stress response elements could be seen across all three analyses, as in the accumulation of osmoprotectant metabolites such as proline and glutamate. Concentrations of potential cell wall precursor amino acids and glucosamine were increased in the VISA strains. Polyamines were decreased in the VISA, which may facilitate the accrual of mutations. Overall, the studies confirm the wide variability in mutations and gene expression patterns that can lead to the VISA phenotype.
Related collections
Most cited references98
- Record: found
- Abstract: found
- Article: not found
Metabolomics--the link between genotypes and phenotypes.
- Record: found
- Abstract: found
- Article: not found
Medium- and short-chain dehydrogenase/reductase gene and protein families
- Record: found
- Abstract: not found
- Article: not found