- Record: found
- Abstract: found
- Article: found
The use of radiocobalt as a label improves imaging of EGFR using DOTA-conjugated Affibody molecule
Read this article at
Abstract
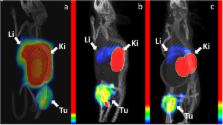
Several anti-cancer therapies target the epidermal growth factor receptor (EGFR). Radionuclide imaging of EGFR expression in tumours may aid in selection of optimal cancer therapy. The 111In-labelled DOTA-conjugated Z EGFR:2377 Affibody molecule was successfully used for imaging of EGFR-expressing xenografts in mice. An optimal combination of radionuclide, chelator and targeting protein may further improve the contrast of radionuclide imaging. The aim of this study was to evaluate the targeting properties of radiocobalt-labelled DOTA-Z EGFR:2377. DOTA-Z EGFR:2377 was labelled with 57Co (T 1/2 = 271.8 d), 55Co (T 1/2 = 17.5 h), and, for comparison, with the positron-emitting radionuclide 68Ga (T 1/2 = 67.6 min) with preserved specificity of binding to EGFR-expressing A431 cells. The long-lived cobalt radioisotope 57Co was used in animal studies. Both 57Co-DOTA-Z EGFR:2377 and 68Ga-DOTA-Z EGFR:2377 demonstrated EGFR-specific accumulation in A431 xenografts and EGFR-expressing tissues in mice. Tumour-to-organ ratios for the radiocobalt-labelled DOTA-Z EGFR:2377 were significantly higher than for the gallium-labelled counterpart already at 3 h after injection. Importantly, 57Co-DOTA-Z EGFR:2377 demonstrated a tumour-to-liver ratio of 3, which is 7-fold higher than the tumour-to-liver ratio for 68Ga-DOTA-Z EGFR:2377. The results of this study suggest that the positron-emitting cobalt isotope 55Co would be an optimal label for DOTA-Z EGFR:2377 and further development should concentrate on this radionuclide as a label.
Related collections
Most cited references46
- Record: found
- Abstract: found
- Article: not found
Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer.
- Record: found
- Abstract: found
- Article: not found
Feedback regulation of EGFR signalling: decision making by early and delayed loops.
- Record: found
- Abstract: found
- Article: not found