- Record: found
- Abstract: found
- Article: found
Detailed interrogation of trypanosome cell biology via differential organelle staining and automated image analysis
Read this article at
Abstract
Background
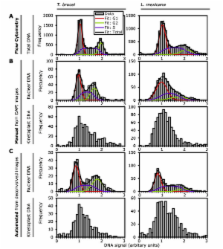
Many trypanosomatid protozoa are important human or animal pathogens. The well defined morphology and precisely choreographed division of trypanosomatid cells makes morphological analysis a powerful tool for analyzing the effect of mutations, chemical insults and changes between lifecycle stages. High-throughput image analysis of micrographs has the potential to accelerate collection of quantitative morphological data. Trypanosomatid cells have two large DNA-containing organelles, the kinetoplast (mitochondrial DNA) and nucleus, which provide useful markers for morphometric analysis; however they need to be accurately identified and often lie in close proximity. This presents a technical challenge. Accurate identification and quantitation of the DNA content of these organelles is a central requirement of any automated analysis method.
Results
We have developed a technique based on double staining of the DNA with a minor groove binding (4'', 6-diamidino-2-phenylindole (DAPI)) and a base pair intercalating (propidium iodide (PI) or SYBR green) fluorescent stain and color deconvolution. This allows the identification of kinetoplast and nuclear DNA in the micrograph based on whether the organelle has DNA with a more A-T or G-C rich composition. Following unambiguous identification of the kinetoplasts and nuclei the resulting images are amenable to quantitative automated analysis of kinetoplast and nucleus number and DNA content. On this foundation we have developed a demonstrative analysis tool capable of measuring kinetoplast and nucleus DNA content, size and position and cell body shape, length and width automatically.
Conclusions
Our approach to DNA staining and automated quantitative analysis of trypanosomatid morphology accelerated analysis of trypanosomatid protozoa. We have validated this approach using Leishmania mexicana, Crithidia fasciculata and wild-type and mutant Trypanosoma brucei. Automated analysis of T. brucei morphology was of comparable quality to manual analysis while being faster and less susceptible to experimentalist bias. The complete data set from each cell and all analysis parameters used can be recorded ensuring repeatability and allowing complete data archiving and reanalysis.
Related collections
Most cited references41
- Record: found
- Abstract: not found
- Article: not found
Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Short communication.
- Record: found
- Abstract: found
- Article: not found