- Record: found
- Abstract: found
- Article: found
Rickettsia monacensis and Human Disease, Spain
brief-report
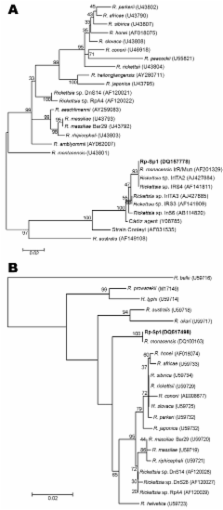
Isabel Jado
* ,
José A. Oteo
† ,
Mikel Aldámiz
‡ ,
Horacio Gil
* ,
Raquel Escudero
* ,
Valvanera Ibarra
† ,
Joseba Portu
‡ ,
Aranzazu Portillo
† ,
María J. Lezaun
‡ ,
Cristina García-Amil
* ,
Isabel Rodríguez-Moreno
* ,
Pedro Anda
*
,
September 2007
Read this article at
There is no author summary for this article yet. Authors can add summaries to their articles on ScienceOpen to make them more accessible to a non-specialist audience.
Abstract
We identified Rickettsia monacensis as a cause of acute tickborne rickettsiosis in 2 humans. Its pathogenic role was assessed by culture and detection of the organism in patients’ blood samples. This finding increases the number of recognized human rickettsial pathogens and expands the known geographic distribution of Mediterranean spotted fever–like cases.
Related collections
Most cited references14
- Record: found
- Abstract: found
- Article: not found
Genotypic identification of rickettsiae and estimation of intraspecies sequence divergence for portions of two rickettsial genes.
R Regnery, C L Spruill, B D Plikaytis (1991)
- Record: found
- Abstract: found
- Article: not found
Rickettsia monacensis sp. nov., a spotted fever group Rickettsia, from ticks (Ixodes ricinus) collected in a European city park.
Jason Simser, Ann Palmer, Volker Fingerle … (2002)
- Record: found
- Abstract: found
- Article: not found
Phylogenetic analysis of spotted fever group rickettsiae based on gltA, 17-kDa, and rOmpA genes amplified by nested PCR from ticks in Japan.
Mitsuhiro Ishikura, Shuji Ando, Yasuhiro Shinagawa … (2002)