- Record: found
- Abstract: found
- Article: found
Role of gut microbiota in type 2 diabetes pathophysiology
Read this article at
Abstract
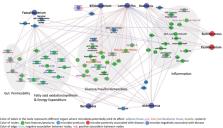
A substantial body of literature has provided evidence for the role of gut microbiota in metabolic diseases including type 2 diabetes. However, reports vary regarding the association of particular taxonomic groups with disease. In this systematic review, we focused on the potential role of different bacterial taxa affecting diabetes. We have summarized evidence from 42 human studies reporting microbial associations with disease, and have identified supporting preclinical studies or clinical trials using treatments with probiotics. Among the commonly reported findings, the genera of Bifidobacterium, Bacteroides, Faecalibacterium, Akkermansia and Roseburia were negatively associated with T2D, while the genera of Ruminococcus, Fusobacterium, and Blautia were positively associated with T2D. We also discussed potential molecular mechanisms of microbiota effects in the onset and progression of T2D.
Related collections
Most cited references78

- Record: found
- Abstract: found
- Article: found
Akkermansia muciniphila -derived extracellular vesicles influence gut permeability through the regulation of tight junctions
- Record: found
- Abstract: not found
- Article: not found
Bacteroides vulgatus and Bacteroides dorei Reduce Gut Microbial Lipopolysaccharide Production and Inhibit Atherosclerosis
- Record: found
- Abstract: found
- Article: not found