- Record: found
- Abstract: found
- Article: found
Morpho-molecular identification and first report of Fusarium equiseti in causing chilli wilt from Kashmir (Northern Himalayas)
Read this article at
Abstract
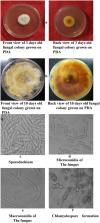
Chilli ( Capsicum annuum L.) is one of the most significant vegetable and spice crop. Wilt caused by Fusarium Sp. has emerged as a serious problem in chilli production. Internal transcribed spacer (ITS) region is widely used as a DNA barcoding marker to characterize the diversity and composition of Fusarium communities. ITS regions are heavily used in both molecular methods and ecological studies of fungi, because of its high degree of interspecific variability, conserved primer sites and multiple copy nature in the genome. In the present study we focused on morphological and molecular characterization of pathogen causing chilli wilt. Chilli plants were collected from four districts of Kashmir valley of Himalayan region. Pathogens were isolated from infected root and stem of the plants. Isolated pathogens were subjected to DNA extraction and PCR amplification. The amplified product was sequenced and three different wilt causing fungal isolates were obtained which are reported in the current investigation. In addition to Fusarium oxysporum and Fusarium solani, a new fungal species was found in association with the chilli wilt in Kashmir valley viz., Fusarium equiseti that has never been reported before from this region. The studies were confirmed by pathogenicity test and re-confirmation by DNA barcoding.
Related collections
Most cited references39
- Record: found
- Abstract: found
- Article: not found
MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets.
- Record: found
- Abstract: found
- Article: not found
Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi.
- Record: found
- Abstract: found
- Article: not found