- Record: found
- Abstract: found
- Article: found
Subcellular Location of PKA Controls Striatal Plasticity: Stochastic Simulations in Spiny Dendrites
Read this article at
Abstract
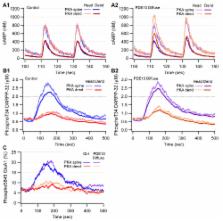
Dopamine release in the striatum has been implicated in various forms of reward dependent learning. Dopamine leads to production of cAMP and activation of protein kinase A (PKA), which are involved in striatal synaptic plasticity and learning. PKA and its protein targets are not diffusely located throughout the neuron, but are confined to various subcellular compartments by anchoring molecules such as A-Kinase Anchoring Proteins (AKAPs). Experiments have shown that blocking the interaction of PKA with AKAPs disrupts its subcellular location and prevents LTP in the hippocampus and striatum; however, these experiments have not revealed whether the critical function of anchoring is to locate PKA near the cAMP that activates it or near its targets, such as AMPA receptors located in the post-synaptic density. We have developed a large scale stochastic reaction-diffusion model of signaling pathways in a medium spiny projection neuron dendrite with spines, based on published biochemical measurements, to investigate this question and to evaluate whether dopamine signaling exhibits spatial specificity post-synaptically. The model was stimulated with dopamine pulses mimicking those recorded in response to reward. Simulations show that PKA colocalization with adenylate cyclase, either in the spine head or in the dendrite, leads to greater phosphorylation of DARPP-32 Thr34 and AMPA receptor GluA1 Ser845 than when PKA is anchored away from adenylate cyclase. Simulations further demonstrate that though cAMP exhibits a strong spatial gradient, diffusible DARPP-32 facilitates the spread of PKA activity, suggesting that additional inactivation mechanisms are required to produce spatial specificity of PKA activity.
Author Summary
The striatum is a part of the basal ganglia which plays a role in addiction and reward learning. Its importance is underscored by pathologies such as Parkinson's disease and Huntington's disease in which degeneration of the dopamine inputs to the striatum or degeneration of neurons in the striatum, respectively, produces motor dysfunction. Dopamine in the striatum activates cascades of signaling molecules, ultimately producing an activity dependent change in the strength of connections between neurons. However, the dispersive movement of signaling molecules seems incompatible with the strengthening of specific subsets of connections, which is required for formation of distinct memories. Anchoring proteins, which restrict molecules to particular compartments within the neuron, are proposed to achieve specificity. We develop a reaction-diffusion model of dopamine activated signaling pathways to explore mechanisms whereby anchoring proteins can produce specificity. We use an efficient Monte-Carlo simulator to implement the cascades of signaling molecules in a neuronal dendrite with multiple dendritic spines. Simulations demonstrate that spatial specificity requires both anchoring proteins and inactivation mechanisms that limit the diffusion of signaling molecules.
Related collections
Most cited references96
- Record: found
- Abstract: found
- Article: not found
AKAP signalling complexes: focal points in space and time.
- Record: found
- Abstract: found
- Article: not found
Regulation of distinct AMPA receptor phosphorylation sites during bidirectional synaptic plasticity.
- Record: found
- Abstract: found
- Article: not found