- Record: found
- Abstract: found
- Article: found
SILGGM: An extensive R package for efficient statistical inference in large-scale gene networks
Read this article at
Abstract
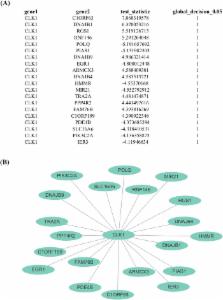
Gene co-expression network analysis is extremely useful in interpreting a complex biological process. The recent droplet-based single-cell technology is able to generate much larger gene expression data routinely with thousands of samples and tens of thousands of genes. To analyze such a large-scale gene-gene network, remarkable progress has been made in rigorous statistical inference of high-dimensional Gaussian graphical model (GGM). These approaches provide a formal confidence interval or a p-value rather than only a single point estimator for conditional dependence of a gene pair and are more desirable for identifying reliable gene networks. To promote their widespread use, we herein introduce an extensive and efficient R package named SILGGM ( Statistical Inference of Large-scale Gaussian Graphical Model) that includes four main approaches in statistical inference of high-dimensional GGM. Unlike the existing tools, SILGGM provides statistically efficient inference on both individual gene pair and whole-scale gene pairs. It has a novel and consistent false discovery rate (FDR) procedure in all four methodologies. Based on the user-friendly design, it provides outputs compatible with multiple platforms for interactive network visualization. Furthermore, comparisons in simulation illustrate that SILGGM can accelerate the existing MATLAB implementation to several orders of magnitudes and further improve the speed of the already very efficient R package FastGGM. Testing results from the simulated data confirm the validity of all the approaches in SILGGM even in a very large-scale setting with the number of variables or genes to a ten thousand level. We have also applied our package to a novel single-cell RNA-seq data set with pan T cells. The results show that the approaches in SILGGM significantly outperform the conventional ones in a biological sense. The package is freely available via CRAN at https://cran.r-project.org/package=SILGGM.
Related collections
Most cited references32
- Record: found
- Abstract: found
- Article: not found
Sparse inverse covariance estimation with the graphical lasso.
- Record: found
- Abstract: found
- Article: not found
A gene-coexpression network for global discovery of conserved genetic modules.
- Record: found
- Abstract: found
- Article: not found