- Record: found
- Abstract: found
- Article: found
A Formal Re-Description of the Cockroach Hebardina concinna Anchored on DNA Barcodes Confirms Wing Polymorphism and Identifies Morphological Characters for Field Identification
Read this article at
Abstract
Background
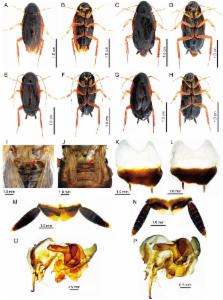
Hebardina concinna is a domestic pest and potential vector of pathogens throughout East and Southeast Asia, yet identification of this species has been difficult due to a lack of diagnostic morphological characters, and to uncertainty in the relationship between macroptyrous (long-winged) and brachypterous (small-winged) morphotypes. In insects male genital structures are typically species-specific and are frequently used to identify species. However, male genital structures in H. concinna had not previously been described, in part due to difficulty in identifying conspecifics.
Methods/Principal Findings
We collected 15 putative H. concinna individuals, from Chinese populations, of both wing morphotypes and both sexes and then generated mitochondrial COI (the standard barcode region) and COII sequences from five of these individuals. These confirmed that both morphotypes of both sexes are the same species. We then dissected male genitalia and compared genital structures from macropterous and brachypterous individuals, which we showed to be identical, and present here for the first time a detailed description of H. concinna male genital structures. We also present a complete re-description of the morphological characters of this species, including both wing morphs.
Conclusions/Significance
This work describes a practical application of DNA barcoding to confirm that putatively polymorphic insects are conspecific and then to identify species-specific characters that can be used in the field to identify individuals and to obviate the delay and cost of returning samples to a laboratory for DNA sequencing.
Related collections
Most cited references8

- Record: found
- Abstract: found
- Article: found
DNA Barcoding the Geometrid Fauna of Bavaria (Lepidoptera): Successes, Surprises, and Questions

- Record: found
- Abstract: found
- Article: found
Design and applicability of DNA arrays and DNA barcodes in biodiversity monitoring
- Record: found
- Abstract: found
- Article: not found