- Record: found
- Abstract: found
- Article: found
Environmental DNA sampling reveals high occupancy rates of invasive Burmese pythons at wading bird breeding aggregations in the central Everglades
Read this article at
Abstract
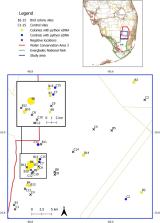
The Burmese python ( Python bivittatus) is now established as a breeding population throughout south Florida, USA. However, the extent of the invasion, and the ecological impacts of this novel apex predator on animal communities are incompletely known, in large part because Burmese pythons (hereafter “pythons”) are extremely cryptic and there has been no efficient way to detect them. Pythons are recently confirmed nest predators of long-legged wading bird breeding colonies (orders Ciconiiformes and Pelecaniformes). Pythons can consume large quantities of prey and may not be recognized as predators by wading birds, therefore they could be a particular threat to colonies. To quantify python occupancy rates at tree islands where wading birds breed, we utilized environmental DNA (eDNA) analysis—a genetic tool which detects shed DNA in water samples and provides high detection probabilities. We fitted multi-scale Bayesian occupancy models to test the prediction that pythons occupy islands with wading bird colonies at higher rates compared to representative control islands containing no breeding birds. Our results suggest that pythons are widely distributed across the central Everglades in proximity to active wading bird colonies. In support of our prediction that pythons are attracted to colonies, site-level python eDNA occupancy rates were higher at wading bird colonies (ψ = 0.88, 95% credible interval [0.59–1.00]) than at the control islands (ψ = 0.42 [0.16–0.80]) in April through June (n = 15 colony-control pairs). We found our water temperature proxy (time of day) to be informative of detection probability, in accordance with other studies demonstrating an effect of temperature on eDNA degradation in occupied samples. Individual sample concentrations ranged from 0.26 to 38.29 copies/μL and we generally detected higher concentrations of python eDNA in colony sites. Continued monitoring of wading bird colonies is warranted to determine the effect pythons are having on populations and investigate putative management activities.
Related collections
Most cited references68
- Record: found
- Abstract: found
- Article: not found
Environmental DNA for wildlife biology and biodiversity monitoring.
- Record: found
- Abstract: not found
- Article: not found
Environmental DNA – An emerging tool in conservation for monitoring past and present biodiversity
- Record: found
- Abstract: found
- Article: not found