- Record: found
- Abstract: found
- Article: not found
Sox4 mediates Tbx3 transcriptional regulation of the gap junction protein Cx43
Read this article at
Abstract
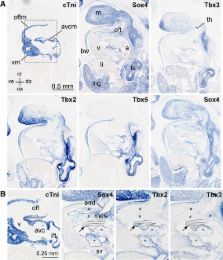
Tbx3, a T-box transcription factor, regulates key steps in development of the heart and other organ systems. Here, we identify Sox4 as an interacting partner of Tbx3. Pull-down and nuclear retention assays verify this interaction and in situ hybridization reveals Tbx3 and Sox4 to co-localize extensively in the embryo including the atrioventricular and outflow tract cushion mesenchyme and a small area of interventricular myocardium. Tbx3, SOX4, and SOX2 ChIP data, identify a region in intron 1 of Gja1 bound by all tree proteins and subsequent ChIP experiments verify that this sequence is bound, in vivo, in the developing heart. In a luciferase reporter assay, this element displays a synergistic antagonistic response to co-transfection of Tbx3 and Sox4 and in vivo, in zebrafish, drives expression of a reporter in the heart, confirming its function as a cardiac enhancer. Mechanistically, we postulate that Sox4 is a mediator of Tbx3 transcriptional activity.
Related collections
Most cited references55
- Record: found
- Abstract: found
- Article: not found
ChIP-seq accurately predicts tissue-specific activity of enhancers.

- Record: found
- Abstract: found
- Article: found
CDD: specific functional annotation with the Conserved Domain Database
- Record: found
- Abstract: found
- Article: not found
