- Record: found
- Abstract: found
- Article: found
The Interleukin-1 (IL-1) Superfamily Cytokines and Their Single Nucleotide Polymorphisms (SNPs)
Read this article at
Abstract
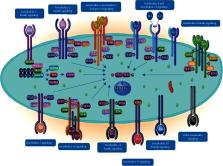
Interleukins (ILs)—which are important members of cytokines—consist of a vast group of molecules, including a wide range of immune mediators that contribute to the immunological responses of many cells and tissues. ILs are immune-glycoproteins, which directly contribute to the growth, activation, adhesion, differentiation, migration, proliferation, and maturation of immune cells; and subsequently, they are involved in the pro and anti-inflammatory responses of the body, by their interaction with a wide range of receptors. Due to the importance of immune system in different organisms, the genes belonging to immune elements, such as ILs, have been studied vigorously. The results of recent investigations showed that the genes pertaining to the immune system undergo progressive evolution with a constant rate. The occurrence of any mutation or polymorphism in IL genes may result in substantial changes in their biology and function and may be associated with a wide range of diseases and disorders. Among these abnormalities, single nucleotide polymorphisms (SNPs) can represent as important disruptive factors. The present review aims at concisely summarizing the current knowledge available on the occurrence, properties, role, and biological consequences of SNPs within the IL-1 family members.
Related collections
Most cited references328
- Record: found
- Abstract: found
- Article: not found
Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics.
- Record: found
- Abstract: found
- Article: not found
Defining trained immunity and its role in health and disease
- Record: found
- Abstract: found
- Article: not found