- Record: found
- Abstract: found
- Article: found
Relationship between intact HIV-1 proviruses in circulating CD4 + T cells and rebound viruses emerging during treatment interruption
Read this article at
Significance
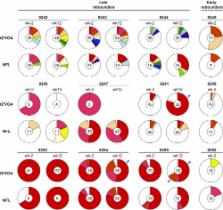
The HIV-1 latent reservoir is the major barrier to cure. Analysis of the replication competent latent reservoir that can be induced in viral outgrowth assays (VOAs) showed little or no overlap with HIV viruses that emerge in plasma after treatment interruption. To determine whether intact proviruses amplified from DNA are more closely related to rebound viruses than those obtained from VOA, we sequenced HIV proviral genomes from CD4 + T cells of individuals who underwent analytical treatment interruption. We find that intact proviruses obtained from DNA overlap in part with those obtained by VOA, but do not overlap with rebound viruses. However, nearly half of all rebound sequences could be accounted for in part by recombination of intact near full-length sequences.
Abstract
Combination antiretroviral therapy controls but does not cure HIV-1 infection because a small fraction of cells harbor latent viruses that can produce rebound viremia when therapy is interrupted. The circulating latent virus reservoir has been documented by a variety of methods, most prominently by viral outgrowth assays (VOAs) in which CD4 + T cells are activated to produce virus in vitro, or more recently by amplifying proviral near full-length (NFL) sequences from DNA. Analysis of samples obtained in clinical studies in which individuals underwent analytical treatment interruption (ATI), showed little if any overlap between circulating latent viruses obtained from outgrowth cultures and rebound viruses from plasma. To determine whether intact proviruses amplified from DNA are more closely related to rebound viruses than those obtained from VOAs, we assayed 12 individuals who underwent ATI after infusion of a combination of two monoclonal anti–HIV-1 antibodies. A total of 435 intact proviruses obtained by NFL sequencing were compared with 650 latent viruses from VOAs and 246 plasma rebound viruses. Although, intact NFL and outgrowth culture sequences showed similar levels of stability and diversity with 39% overlap, the size of the reservoir estimated from NFL sequencing was larger than and did not correlate with VOAs. Finally, intact proviruses documented by NFL sequencing showed no sequence overlap with rebound viruses; however, they appear to contribute to recombinant viruses found in plasma during rebound.
Related collections
Most cited references18
- Record: found
- Abstract: found
- Article: not found
Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy.
- Record: found
- Abstract: found
- Article: not found
Persistence of HIV in gut-associated lymphoid tissue despite long-term antiretroviral therapy.
- Record: found
- Abstract: found
- Article: not found