- Record: found
- Abstract: found
- Article: found
Innate Sensing of Chitin and Chitosan
research-article
10 January 2013
Read this article at
There is no author summary for this article yet. Authors can add summaries to their articles on ScienceOpen to make them more accessible to a non-specialist audience.
Abstract
Chitin is the second most common polysaccharide found in nature. It is present in
crustacean shells, insect exoskeletons, parasitic nematode eggs and gut linings, and
in the cell wall of fungi. The deacetylated derivative of chitin, chitosan, is less
common but is particularly evident in certain species of fungi, such as Cryptococcus,
and the cyst wall of Entamoeba. How mammals sense and respond to these polymers is
not well understood, and conflicting reports on their immunological activity have
led to some controversy. Despite this, promising translational applications that exploit
the unique properties of chitin and chitosan are being developed.
What Are Chitin and Chitosan?
Chitin, a linear, neutrally charged polymer of β-(1,4)-linked N-acetylglucosamine
(GlcNAc), and its deacetylated derivative chitosan, a cationic polymer of glucosamine
(GlcN), are two naturally occurring polysaccharides (Figure 1). Using cytoplasmic
stores of UDP-GlcNAc, chitin synthases (EC 2.4.1.16) extrude chitin through the plasma
membrane to an extracellular location [1]. Chitin deacetylases (EC 3.5.1.41), if present,
remove the acetyl group following extrusion. During synthesis, chitin polymers anneal
to one another, typically in opposite orientation (α-chitin), to form fibers of high
tensile strength [2]. The fibers are cross-linked with glucans in fungi to form a
meshwork reinforcing the cell wall and with protein in insect exoskeletons to give
an ordered, laminate structure to the cuticle. Chitinases (EC 3.2.1.14) and chitosanases
(EC 3.2.1.132) secreted by bacteria and fungi recycle gigatons of chitin/chitosan
to GlcNAc/GlcN annually. In their more specialized roles, chitinases are important
in developmental processes, such as remodeling the fungal cell wall and shedding old
cuticle (molting) by crustaceans. For organisms that do not synthesize chitin, plant
and mammalian chitinases aid in defense against chitin-bearing pathogens.
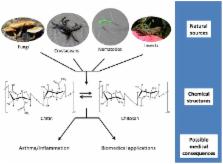
10.1371/journal.ppat.1003080.g001
Figure 1
Chitin and chitosan: from source to consequence.
Chitin and chitosan are naturally found in fungal cell walls, crustacean shells, nematodes
eggs and gut linings, and insect exoskeletons. These polymers consist of long chains
of N-acetylglucosamine (chitin) or glucosamine (chitosan). Conversion between the
two polysaccharides can be performed chemically or happen within the organisms via
chitin deacetylases. Mammalian exposure to the polymers has been linked to both upregulation
and downregulation of inflammatory responses, including those involved in asthma.
Despite this, chitin and chitosan are being utilized in a variety of biomedical applications,
including tissue engineering and drug delivery.
Commercial preparations of the polymers typically begin with isolation of chitin from
crustacean shell waste, followed by chemical deacetylation to chitosan using high
temperature and strong alkali. Particulate crystals of chitin are insoluble while
chitosan is soluble in dilute acid. Commercial preparations of both polymers vary
with regard to degrees of acetylation, polymer lengths, particle size, and contamination
[3], [4]. Such heterogeneity likely accounts for some of the conflicting results in
the literature.
How Are Chitin and Chitosan Recognized by Mammals?
The lack of chitin or chitosan in mammalian cells makes these polymers potential targets
for recognition by the innate immune system. Though chitin and possibly chitosan in
their native environment can be stained with low molecular weight dyes, they are not
readily accessed by protein-sized probes. Their exposure requires some degree of degeneration
of their surrounding architecture, as occurs in damaged cells or fungal/crustacean
detritus [3]. Upon exposure chitin can be recognized by mammalian chitinases, which
bind and actively degrade chitin, and chitinase-like proteins, which also bind chitin
but are catalytically inactive. It has become evident that this family of chitin-binding
proteins (Glycosyl Hydrolase Family 18) plays an active role in inflammation and innate
and adaptive immunity based on their upregulation during various disease states [5].
Both chitin and chitosan particles are readily phagocytosed [4], supporting a role
for recognition via specific receptor(s) mediating phagocytosis. Receptors on myeloid
cells that bind chitin or chitosan and induce a phagocytic response have yet to be
definitively identified. However, several receptors have been shown to have an affinity
for chitin or chitin oligosaccharides, including: FIBCD1, a homotetrameric 55-kDa
type II transmembrane protein expressed in the gastrointestinal tract [6]; NKR-P1,
an activating receptor on rat natural killer cells [7]; RegIIIγ, a secreted C-type
lectin [8]; and galectin-3, a lectin with affinity for β-galactosides [9]. However,
none of these has yet been shown to act as a receptor as opposed to a protein that
binds chitin. Also, receptors that recognize soluble oligosaccharides as by-products
of chitinase digestion may not recognize full-length, insoluble chitin.
What Kind of Responses Does Chitin Elicit in Mammals?
Exposure to chitin, either through food or inhalation, is common. Chitin has been
shown to induce a response similar to the response generated in helminth and allergic
immunity, with an accumulation of eosinophils and basophils expressing IL-4, and alternatively
activated macrophages [10]. Conversely, chitin downregulated the allergic response
to ragweed in mice [11]. Also, asthma/allergic conditions feature alternatively activated
macrophages, which have high expression levels of chitinases and chitinase-like proteins.
Blocking acidic mammalian chitinase (AMCase) or knocking out BRP-39 (chitinase-like
protein) results in decreased inflammation and eosinophilia [12].
Three innate immune receptors, Toll-like receptor (TLR) 2, Dectin-1, and the mannose
receptor, have been implicated in mediating a variety of immune responses to chitin.
However, how this occurs is not well understood. Direct binding to chitin has not
been demonstrated, and the possibility that contaminants are responsible for some
of these effects cannot be excluded. One study showed chitin acting via an apparent
Dectin-1 dependent, but mincle (a C-type lectin), TLR2, and TLR4-independent mechanism
could partially block cytokine production in response to Candida albicans
[3]. Nevertheless, chitin was not shown to directly interact with Dectin-1. However,
TLR2 was found to contribute to sensing of chitin by keratinocytes [13] and chitin-induced
expression of IL-17A and IL-17AR [14]. Moreover, TNFα and IL-10 induced by chitin
appeared to be mediated by TLR2, Dectin-1, and the mannose receptor. Interestingly,
the size of the chitin particles determined the type of response observed: smaller
fragments (<40 µm) induced cytokines that inhibited tissue inflammation, modest-sized
fragments (40–70 µm) induced a strong pro-inflammatory response, and larger fragments
were relatively inert [15]. Finally, though chitin preparations of varying sizes did
not stimulate IL-1β production, chitosan was shown to activate the NLRP3 inflammasome,
leading to robust IL-1β responses by a phagocytosis-dependent mechanism [4].
How Do Plants Recognize and Respond to Chitin and Chitosan?
Fungi are major crop pathogens. It is not surprising then that plants exhibit a wide
variety of defense responses to chitin and chitosan following fungal infestation,
including increases in chitinase expression, proteinase inhibitors, reactive oxygen
species (ROS), cytoplasmic acidification, and expression of early responsive genes
and defense genes [16]. Presumably, most of these responses have developed to fight
fungal infections, though chitin-binding lectins have also been shown to have insecticidal
activity [17]. Likewise, fungi have developed methods to avoid recognition of chitin
and thereby prevent the effective antifungal response, such as masking chitin with
α-1,3-glucan, a compound plants are unable to digest [18]. Conversely, recognition
of modified chitin oligosaccharides is important for symbiotic relationships between
leguminous plants and rhizobial bacteria [19].
A number of receptors in plants that bind directly to chitin or mediate the response
to chitin have been identified. Chitin elicitor-binding proteins (CEBiP) containing
an extracellular lysin motif (LysM) that binds chitin directly are conserved across
multiple plant species. CEBiP knockdown in suspension-cultured rice cells results
in an absence of ROS generation in response to chitin [20]. CERK1, the Arabidopsis
CEBiP homolog, is essential for chitin elicitor signaling; dimerization upon binding
is critical for MAPK activation, ROS generation, and gene expression in response to
chitin [21]. In contrast, chitosan appears to elicit activity from plant cells via
charge-charge interactions with negatively charged phospholipids instead of via a
receptor-specific interaction [22]. Whether analogous charge-based, receptor-free
interactions between mammalian cells and this highly positively charged polymer occur
is speculative.
How Are These Polymers Being Used Translationally?
The unique structural and biological properties of chitin and chitosan are increasingly
being exploited for use in biomedical applications, such as tissue scaffolds and wound
dressings. This has been facilitated by advances in technology to produce purified
polymers with desired physical properties. For example, particle size can be manipulated
to control the resulting inflammatory response. The polycationic properties of chitosan
are being developed for use in biosensors by immobilizing enzymes, in wound dressings
to induce cell migration and proliferation at the wound site, and in tissue engineering
as a scaffold [23].
The polycationic and biodegradable properties of chitosan make it attractive as a
controlled delivery system for conjugated materials. Mucosal vaccines adjuvanted with
chitosan have elicited robust antibody and T-cell responses [24]. Similarly, chitosan
has been shown to have potential utility as a delivery system for drugs and genes
[25]. Although there appears to be promising future applications for these polymers,
currently chitin and chitosan are approved by the US Food and Drug Administration
only for use as food additives. However, there are a number of ongoing clinical trials
looking to expand their approved roles.
Conclusions
Recent research has begun to clarify when and how mammals and plants recognize and
respond to exposure to chitin and chitosan. Nevertheless, there are still many unanswered
questions. Disparities in the literature regarding the immunological activity of chitin
and chitosan are likely due in large part to the relative purity and heterogeneity
of the glycan preparations used as stimuli. In particular, recent studies have demonstrated
an inverse relationship between particle size and immunological activity. While much
progress has been made in elucidating how plants recognize chitin and chitosan, the
principal receptor(s) responsible for mammalian recognition remain to be determined.
Finally, the biodegradable and physicochemical properties of chitin and chitosan make
these glycans ideal for a wide range of translational applications.
Related collections
Most cited references18
- Record: found
- Abstract: found
- Article: not found
Symbiotic bacteria direct expression of an intestinal bactericidal lectin.
Heather L Cash, Cecilia V. Whitham, Cassie Behrendt … (2006)
- Record: found
- Abstract: found
- Article: not found
CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis.
Ayako Miya, Premkumar Albert, Tomonori Shinya … (2007)
- Record: found
- Abstract: found
- Article: not found
Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor.
Hanae Kaku, Yoko Nishizawa, Naoko Ishii-Minami … (2006)