- Record: found
- Abstract: found
- Article: found
RNA-seq analysis reveals significant transcriptome changes in turbot ( Scophthalmus maximus) suffering severe enteromyxosis
Read this article at
Abstract
Background
Enteromyxosis caused by the intestinal myxozoan parasite Enteromyxum scophthalmi is a serious threat for turbot ( Scophthalmus maximus, L.) aquaculture, causing severe catarrhal enteritis leading to a cachectic syndrome, with no therapeutic options available. There are still many aspects of host-parasite interaction and disease pathogenesis that are yet to be elucidated, and to date, no analysis of the transcriptomic changes induced by E. scophthalmi in turbot organs has been conducted. In this study, RNA-seq technology was applied to head kidney, spleen and pyloric caeca of severely infected turbot with the aim of furthering our understanding of the pathogenetic mechanisms and turbot immune response against enteromyxosis.
Results
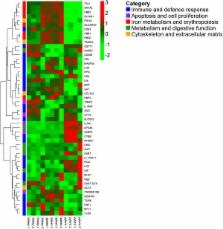
A huge amount of information was generated with more than 23,000 identified genes in the three organs, amongst which 4,762 were differently expressed (DE) between infected and control fish. Associate gene functions were studied based on gene ontology terms and available literature, and the most interesting DE genes were classified into five categories: 1) immune and defence response; 2) apoptosis and cell proliferation; 3) iron metabolism and erythropoiesis; 4) cytoskeleton and extracellular matrix and 5) metabolism and digestive function. The analysis of down-regulated genes of the first category revealed evidences of a connexion failure between innate and adaptive immune response, especially represented by a high number of DE interferon-related genes in the three organs. Furthermore, we found an intense activation of local immune response at intestinal level that appeared exacerbated, whereas in kidney and spleen genes involved in adaptive immune response were mainly down-regulated. The apoptotic machinery was only clearly activated in pyloric caeca, while kidney and spleen showed a marked depression of genes related to erythropoiesis, probably related to disorders in iron homeostasis. The genetic signature of the causes and consequences of cachexia was also demonstrated by the down-regulation of the genes encoding structural proteins and those involved in the digestive metabolism.
Related collections
Most cited references94
- Record: found
- Abstract: found
- Article: not found
Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3 receptor that supports primary immune responses.
- Record: found
- Abstract: found
- Article: not found
Increased expression of interleukin 17 in inflammatory bowel disease.
- Record: found
- Abstract: found
- Article: not found