- Record: found
- Abstract: found
- Article: found
Genome-Wide Detection of Gene Coexpression Domains Showing Linkage to Regions Enriched with Polymorphic Retrotransposons in Recombinant Inbred Mouse Strains
Abstract
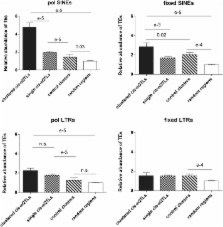
Although gene coexpression domains have been reported in most eukaryotic organisms, data available to date suggest that coexpression rarely concerns more than doublets or triplets of adjacent genes in mammals. Using expression data from hearts of mice from the panel of AxB/BxA recombinant inbred mice, we detected (according to window sizes) 42−53 loci linked to the expression levels of clusters of three or more neighboring genes. These loci thus formed “ cis-expression quantitative trait loci (eQTL) clusters” because their position matched that of the genes whose expression was linked to the loci. Compared with matching control regions, genes contained within cis-eQTL clusters showed much greater levels of coexpression. Corresponding regions showed: (1) a greater abundance of polymorphic elements (mostly short interspersed element retrotransposons), and (2) significant enrichment for the motifs of binding sites for various transcription factors, with binding sites for the chromatin-organizing CCCTC-binding factor showing the greatest levels of enrichment in polymorphic short interspersed elements. Similar cis-eQTL clusters also were detected when we used data obtained with several tissues from BxD recombinant inbred mice. In addition to strengthening the evidence for gene expression domains in mammalian genomes, our data suggest a possible mechanism whereby noncoding polymorphisms could affect the coordinate expression of several neighboring genes.
Most cited references34
- Record: found
- Abstract: found
- Article: not found
Genetics of gene expression surveyed in maize, mouse and man.
- Record: found
- Abstract: found
- Article: not found
Genomic Views of Distant-Acting Enhancers
- Record: found
- Abstract: found
- Article: not found