- Record: found
- Abstract: found
- Article: found
Annotation of the Protein Coding Regions of the Equine Genome

Read this article at
Abstract
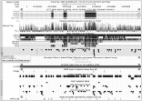
Current gene annotation of the horse genome is largely derived from in silico predictions and cross-species alignments. Only a small number of genes are annotated based on equine EST and mRNA sequences. To expand the number of equine genes annotated from equine experimental evidence, we sequenced mRNA from a pool of forty-three different tissues. From these, we derived the structures of 68,594 transcripts. In addition, we identified 301,829 positions with SNPs or small indels within these transcripts relative to EquCab2. Interestingly, 780 variants extend the open reading frame of the transcript and appear to be small errors in the equine reference genome, since they are also identified as homozygous variants by genomic DNA resequencing of the reference horse. Taken together, we provide a resource of equine mRNA structures and protein coding variants that will enhance equine and cross-species transcriptional and genomic comparisons.
Related collections
Most cited references11
- Record: found
- Abstract: found
- Article: not found
Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction.
- Record: found
- Abstract: found
- Article: found
The UCSC Genome Browser database: update 2011
- Record: found
- Abstract: found
- Article: not found