- Record: found
- Abstract: found
- Article: not found
Genome-Wide Analysis of Factors Affecting Transcription Elongation and DNA Repair: A New Role for PAF and Ccr4-Not in Transcription-Coupled Repair
Read this article at
Abstract
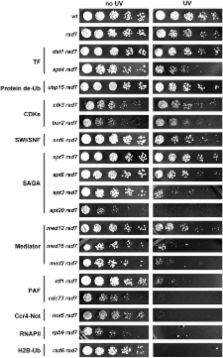
RNA polymerases frequently deal with a number of obstacles during transcription elongation that need to be removed for transcription resumption. One important type of hindrance consists of DNA lesions, which are removed by transcription-coupled repair (TC-NER), a specific sub-pathway of nucleotide excision repair. To improve our knowledge of transcription elongation and its coupling to TC-NER, we used the yeast library of non-essential knock-out mutations to screen for genes conferring resistance to the transcription-elongation inhibitor mycophenolic acid and the DNA-damaging agent 4-nitroquinoline-N-oxide. Our data provide evidence that subunits of the SAGA and Ccr4-Not complexes, Mediator, Bre1, Bur2, and Fun12 affect transcription elongation to different extents. Given the dependency of TC-NER on RNA Polymerase II transcription and the fact that the few proteins known to be involved in TC-NER are related to transcription, we performed an in-depth TC-NER analysis of a selection of mutants. We found that mutants of the PAF and Ccr4-Not complexes are impaired in TC-NER. This study provides evidence that PAF and Ccr4-Not are required for efficient TC-NER in yeast, unraveling a novel function for these transcription complexes and opening new perspectives for the understanding of TC-NER and its functional interconnection with transcription elongation.
Author Summary
Dealing with DNA lesions is one of the most important tasks of both prokaryotic and eukaryotic cells. This is particularly relevant for damage occurring inside genes, in the DNA strands that are actively transcribed, because transcription cannot proceed through a damaged site and the persisting lesion can cause either genome instability or cell death. Cells have evolved specific mechanisms to repair these DNA lesions, the malfunction of which leads to severe genetic syndromes in humans. Despite many years of intensive research, the mechanisms underlying transcription-coupled repair is still poorly understood. To gain insight into this phenomenon, we undertook a genome-wide screening in the model eukaryotic organism Saccharomyces cerevisiae for genes that affect this type of repair that is coupled to transcription. Our study has permitted us to identify and demonstrate new roles in DNA repair for factors with a previously known function in transcription, opening new perspectives for the understanding of DNA repair and its functional interconnection with transcription.
Related collections
Most cited references76
- Record: found
- Abstract: found
- Article: not found
The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae.
- Record: found
- Abstract: found
- Article: not found
Elongation by RNA polymerase II: the short and long of it.
- Record: found
- Abstract: not found
- Article: not found
