- Record: found
- Abstract: found
- Article: found
Novel regional age-associated DNA methylation changes within human common disease-associated loci

Read this article at
Abstract
Background
Advancing age progressively impacts on risk and severity of chronic disease. It also modifies the epigenome, with changes in DNA methylation, due to both random drift and variation within specific functional loci.
Results
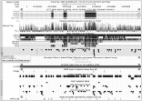
In a discovery set of 2238 peripheral-blood genome-wide DNA methylomes aged 19–82 years, we identify 71 age-associated differentially methylated regions within the linkage disequilibrium blocks of the single nucleotide polymorphisms from the NIH genome-wide association study catalogue. This included 52 novel regions, 29 within loci not covered by 450 k or 27 k Illumina array, and with enrichment for DNase-I Hypersensitivity sites across the full range of tissues. These age-associated differentially methylated regions also show marked enrichment for enhancers and poised promoters across multiple cell types. In a replication set of 2084 DNA methylomes, 95.7 % of the age-associated differentially methylated regions showed the same direction of ageing effect, with 80.3 % and 53.5 % replicated to p < 0.05 and p < 1.85 × 10 –8, respectively.
Related collections
Most cited references45

- Record: found
- Abstract: found
- Article: found
DNA methylation age of blood predicts all-cause mortality in later life
- Record: found
- Abstract: found
- Article: found
The UCSC Genome Browser database: update 2011
- Record: found
- Abstract: not found
- Article: not found