INTRODUCTION
The importance of lipids in biology is without question as they act as secondary messengers and signaling molecules that are added post-translationally to proteins to regulate their function and targeting. Lipids are also responsible for the formation of membrane barriers between cellular compartments. Membrane lipids hold a unique place among cellular biomolecules for their dual nature: They have both hydrophobic long carbon side chains and hydrophilic polar head groups that enable them to interact with both organic and aqueous environments. To form lipid bilayer membranes, the hydrophobic regions interact with one another while their polar side chains on the outside interact with the aqueous cellular environment. At the same time, the lipids at the edge of the cellular compartment provide a stably organized hydrophobic environment to support functions such as assembly and transport of various lipid signaling molecules, lipid-modified proteins, and membrane proteins. Moreover, the interplay between lipids and proteins has been shown to alter the structural properties of proteins [1] and this has significant ramifications for human diseases such as Alzheimer's, Parkinson's, and prion diseases [2]. Both membrane lipids and lipids lacking strong polar groups serve as signaling molecules in apoptosis [3], in lymphocyte activation [4], in membrane targeting inhibition [5], and systemic functions in organisms ranging from inflammation to cancer. Lipid functions as energy storage molecules are well known (e.g. triglycerides) and yet they also serve a wide range of essential functions, such as the source of fat-soluble vitamins and metabolic regulators. Unsurprisingly, lipid metabolism is critical for cell fate as well as for health and disease, being involved in respiratory and renal pathologies, in obesity, in neurological disorders, and finally in a wide range of cardiovascular diseases and cancer [6].
Part of what makes lipids such an effective cellular component is their enormous diversity. There are just four nucleotides that code for the entire human genome and 21 amino acids that code for proteins. On the other hand, there are hundreds and possibly thousands of distinct lipid types in any given cell. This is possible because of the building block nature – even within each individual lipid. Each additional carbon atom in a lipid molecule can change its properties and typical cellular lipids range from 6 to 28 carbons. Similarly, each double bond added within the lipid carbon chain further alters its properties. For example, many microorganisms change their lipid membrane profile with changing temperatures or pressure (such as deep ocean organisms) to increase the number of double bonds in their side chains, thus rendering the membranes more rigid [7,8]. Moreover, the more complex lipids have multiple carbon chains in addition to a variety of polar head groups, which could essentially lead to an infinite number of possibilities.
Lipidomics aims to identify and characterize the lipids of the cell and to further investigate their structure and functions. Lipidomes of various organisms such as fruit fly have been recently reported [9], along with the full-lipidome profiling of yeast [10], blood plasma [11], viruses [12,13], and mammalian epithelial cell lines [14,15]. Whereas the lipid composition of Drosophila melanogaster and Homo sapiens are strikingly alike, in other cases, the abundance of a particular lipid compared to another is directly reversed (see Figure 1). It is reasonable to wonder whether the unchanging lipids perform an indispensable function. On the other hand, it is not always clear whether glaring differences such as the enrichment of phosphatidylinositol (PI) in Saccharomyces cerevisiae plasma and endosomal membranes [16] indicate unique functions for specific needs of different organisms or if these dissimilarities simply reflect divergent evolution.

Although large-scale projects such as Lipid Maps and Eurolipidomics have resulted in significant progress to this field, very little is known about the function of lipids inside the nucleus [17]. Phospholipids (PLs), ceramides (Cer), and sphingosines were found to impact on replication and transcription in both prokaryotes and eukaryotes [18]. For example, gene expression was altered upon treatment of the cells with gamma-linoleic acid [19]. Negatively charged PLs such as PI, cardiolipin (CL), phosphatidylserine (PS), and phosphatidylglycerol (PG) stimulate RNA synthesis, while phosphatidylcholine (PC), phosphatidylethanolamine (PE), and sphingomyelin (SM) inhibit this process [20]. Although the extremely complex lipid environment within a cell often makes it difficult to distinguish direct from indirect effects, clear lipid effects on nuclear functions have also been shown in fully controlled in vitro experiments. For example, a recent study indicated that a particular sphingolipid could modulate DNA polymerase activity [21].
There are many plausible hypothetical mechanisms for lipid nuclear functions beyond membrane compartmentalization and signaling. Lipids could directly affect DNA supercoiling, which is important for transcription, replication, and recombination/repair. Consistent with this hypothesis, selected natural and synthetic lipids inhibit the functioning of prokaryotic topoisomerase and polymerase enzymes via yet unidentified mechanisms [22–24]. Here, we will review some of the nuclear lipids and discuss their known and/or possible functions. In particular, we will describe experiments that led us to hypothesize that lipids may constitute a significant informational layer in the gene regulation space, conceptually partial to, what previously has been referred to as, “epigenetic code.” While we are only starting to isolate lipid moieties, serving as structural constituents of chromatin, it is increasingly clear that the involvement of lipids in gene regulation is manyfold. While emerging/future technologies, such as lipid labeling, lipid–protein and lipid–DNA crosslinking, and super-resolution microscopy, warrant exciting big data on molecular mechanisms that cells employ to engage lipids in order to regulate epigenetic processes, we will only briefly touch upon cases where certain lipids could be placed on the map, for playing roles in gene expression and chromosome dynamics. We will first look at the nuclear envelope, get through to its inner side, then briefly look inside the nucleoplasm, at the chromatin, the “genome wrap,” and in conclusion, at the metaboloepigenetic transactions, dynamically engaging nuclear proteins (including histones) on one side, and lipids on the other.
THE LIPID ENVIRONMENT OF THE NUCLEAR ENVELOPE
The various cell membranes differ in lipid composition. For instance, PC is the most abundant lipid in cell membranes of many species, while sphingolipids are next on the list being observed in the plasma membrane, while they are almost undetectable in the endoplasmic reticulum (ER), where PE and PI considerably contribute [16]. It has also been observed that the lipid composition changes between the ER and Golgi complexes [16]. Indeed, the lipid composition of each cellular organelle has been found to be both unique and highly conserved across evolution, in such a way that only the plasma membrane is distinct in its lipid composition between yeast and man. However, various laboratories trying to determine the lipid composition of the nuclear envelope, even within the same organism, obtained inconsistent results; hence the precise composition of the nuclear envelope remains uncertain. This likely reflects several unique aspects of the nucleus as a membrane bound compartment. First, the nucleus has two distinct lipid bilayers, the inner and outer nuclear membranes, each of which could have a distinct composition. The outer nuclear membrane is physically continuous with the ER and so ER lipids can freely diffuse and mix with it. Nonetheless, proteomics has identified lipid synthesis enzymes in the inner nuclear membrane that could be responsible for generating its distinct composition [25]. Second, the nuclear envelope often has invaginations that contain mitochondria, perhaps to swiftly provide energy for nuclear functions, whose numbers vary enormously. Unfortunately, they are almost impossible to extract while maintaining the nuclear membrane intact. Finally, the nucleus has its own storage of molecules, such as cholesterol and inositol phosphate, which makes it impossible to determine if the isolated cholesterol is intra-nuclear or membrane-associated.
Though the precise lipid composition of the nuclear envelope remains elusive, the protein composition of the nuclear envelope has been comprehensively determined [25–28]. Many proteins involved in lipid synthesis and membrane function were identified based on GO-term associations. It was found that certain types of lipid-related proteins and enzymes were particularly enriched in the nuclear envelopes, compared to their percentage representation against all GO-term-associated proteins encoded in the genome (Table 1). The identification of so many proteins involved in lipid biogenesis argues that during interphase, where nuclear size increases twofold to threefold in S-phase, new lipids are generated inside the nucleus, and not from lateral diffusion from the ER. This further supports the possibility that the lipid composition of the inner nuclear membrane could differ from that of the outer nuclear membrane/ER. Such lipid synthesizing and metabolic proteins in the nuclear envelope could also function to support a myriad of possible lipid modifications on chromatin and their potential roles in epigenetic regulation.
| GO-term | NE proteins | Percentage of NE protein | Percentage of all in genome | P-value |
|---|---|---|---|---|
| Membrane organization | 173 | 7.3 | 3.0 | 5.18e–23 |
| Nucleus organization | 40 | 1.7 | 0.3 | 1.61e–16 |
| Endomembrane system organization | 91 | 3.9 | 1.4 | 1.78e–15 |
| Localization within membrane | 14 | 0.6 | 0.1 | 1.75e–03 |
| Response to topologically incorrect protein | 42 | 1.8 | 0.4 | 7.76e–12 |
| Lipid metabolic process | 172 | 7.3 | 4.0 | 4.27e–11 |
| Neutral lipid metabolic process | 31 | 1.3 | 0.3 | 1.02e–07 |
| Membrane disassembly | 19 | 0.8 | 0.1 | 4.92e–10 |
| Establishment of protein localization to membrane | 59 | 2.5 | 0.9 | 8.34e–10 |
| Lipid catabolic process | 41 | 1.7 | 0.7 | 2.39e–03 |
| Glycero-lipid metabolic process | 55 | 2.3 | 1.1 | 2.94e–03 |
| Arachidonic acid metabolic process | 17 | 0.7 | 0.2 | 9.26e–03 |
| Fatty acid metabolic process | 62 | 2.6 | 0.9 | 2.50e–09 |
| Long-chain fatty acid metabolic process | 24 | 1.0 | 0.3 | 3.4e–04 |
| Very long chain fatty acid metabolic process | 10 | 0.4 | 0.1 | 4.70e–03 |
| Triglyceride metabolic process | 29 | 1.2 | 0.3 | 5.08e–07 |
| Triglyceride biosynthetic process | 17 | 0.7 | 0.2 | 2.8e–04 |
| Fatty acid biosynthetic process | 28 | 1.2 | 0.4 | 7.4e–04 |
| Lipid biosynthetic process | 90 | 3.8 | 1.7 | 8.47e–09 |
| Neutral lipid biosynthetic process | 17 | 0.7 | 0.2 | 4.4e–04 |
| Carbohydrate biosynthetic process | 38 | 1.6 | 0.6 | 3.37e–05 |
| Internal protein amino acid acetylation | 33 | 1.4 | 0.4 | 3.89e–06 |
To see original data created using Termfinder [29] from which Table was generated you can visit: https://github.com/AlastairKerr/Zhdanov-Kagansky.
Hypothesis: Proteins involved in lipid metabolism are strikingly enriched within the nuclear envelope, which could accommodate dynamic changes in the structure of the nucleus throughout the cell cycle without the need to transport lipids through the hydrophilic cytoplasm and this could also support nuclear lipid signaling and regulatory functions.
The degree to which chromatin could come into contact with the nuclear membrane itself is unclear. Electron microscopy shows electron dense material directly apposed to the membrane, but cannot clarify whether the chromatin loops through the lamin polymer were able to directly interact. Certain nuclear envelope transmembrane (NET) proteins [30] interact directly with chromatin, but whether NETs penetrate beyond the lamin polymer for this interaction or the chromatin penetrates the lamin polymer to reach the NET is yet unclear. Lipids may also be able to interact with chromatin during mitosis under two circumstances: i) breaking down of nuclear envelope and ii) reformation of nuclear envelope, when presumably vesicles and tubules containing NETs and associated with chromatin binding proteins assemble around the mitotic chromosomes. Whether the chromatin can directly access the membrane or synthesized lipids are able to covalently attach to chromatin proteins independent of the membrane, lipid interactions could contribute to compact packaging and silencing of chromatin at the nuclear periphery, providing a convenient supplement to the epigenetic control of multi-megabase satellite repeats that account for almost half of the genome in humans [31]. The seemingly surprising fact that lipid synthesizing, modifying, and catabolic enzymes have not yet been identified by functional genomics may be explained by most lipids being indispensable for the cells for their other signaling and structural roles.
LIPID MODIFICATIONS OF NUCLEAR ENVELOPE PROTEINS
Many small GTPases that function at the plasma membrane are anchored to the membrane via a covalently added lipid moiety, typically farnesylation, or geranylgeranylation [32]. Such proteins are moderately abundant but sporadic at the plasma membrane and are primarily involved in signaling pathways. In the nuclear envelope, the intermediate filament lamins are post-translationally farnesylated. In contrast to plasma membrane GTPases, the lamins are one of the most abundant proteins in the nucleus, estimated at up to 9 million copies in a single nucleus [33], and assemble into a polymer lining the inner nuclear membrane. There are three genes encoding lamins (LMNA, LMNB1, LMNB2) and each of them has multiple splice variants. The major forms of all three are farnesylated; however, whereas the farnesylation is permanent for lamins B1 and B2, it is only transient for lamin A [32]. Some studies suggest that the different lamin subtypes form distinct networks that layer with the permanently farnesylated lamins B1 and B2, more proximal to the membrane than lamin A [34,35]. Accordingly, mutation of each yields strikingly distinct phenotypes [36].
The most intriguing diseases linked to lamins include, restrictive dermopathy, and the premature ageing progeroid syndromes including Atypical Werner's Syndrome and Hutchison-Gilford Progeria Syndrome (HGPS) [37,38]. Restrictive dermopathy is embryonic lethal, while the progeroid syndromes are principally characterized by growth retardation, alopecia, facial hypoplasia, premature arteriosclerosis, and osteoporosis. Both syndromes involve a disruption of the normal processing of the lamin A farnesylation. Usually, the farnesyl group is added by a farnesyltransferase to the cystein within a CaaX motif at the C-terminus of the protein. Next, the last three amino acids are cleaved and the cystein is carboxy-methylated. Finally, the last 15 amino acids (so 18 in total) are cleaved, thus releasing the farnesyl group from the processed lamin protein. In the case of restrictive dermopathy, the defect is caused just by the inability to properly remove the C-terminal amino acids along with the farnesyl group. Accordingly, it can also be caused by mutations in the Zmpste24 protease that normally performs the cleavage step. In the case of HGPS, the most common lamin A mutation affects a splice junction so that the exon containing the cleavage site is lost. This results in a permanently farnesylated lamin A protein-like restrictive dermopathy, but it also results in a protein lacking 50 amino acids from the missing exon; so it is more difficult to pin the pathologies on just the lipid moiety. Nonetheless, the farnesyl group has been a principal target of both disorders and treatment with farnesyltransferase inhibitors proven to be very promising in early in vitro studies [39]; however, effects were minimal when trialed in patients [40].
Little is known about whether there are other lipid-modified proteins associated with the inner nuclear membrane or how the presence or absence of the lipid moiety affects lamin polymer assembly as the detailed structure of the lamin polymer itself remains unclear. However, there are not likely to be many other farnesylated proteins, as the CaaX motif is present on few other proteins in the nuclear envelope proteome. Nonetheless, inositol triphosphate signaling has been clearly found in the nuclear envelope [41,42] and probably many more types of lipid signaling molecules will be found when the nuclear envelope lipidome is finally directly investigated.
LIPIDS FOUND INSIDE THE NUCLEOPLASM
Varying lipid content of the nuclei from different cell types was reported almost a century ago [43]. In this study, a relatively high content of lipids, especially PL and cholesterol, was found in nuclei (see Figure 2). The overall concept of epigenetic regulation in metazoans, proposed a few years later, was successfully then used to explain drastic phenotypic diversification in development between cells in different tissues as well as in disease [44]. The proteins identified in the nuclear envelope, many of which are highly tissue-specific [25–28], raise the possibility of differences in lipid composition of the inner nuclear membrane between tissues. Such differences, if confirmed experimentally in the nuclear envelope lipidome, could be important for epigenetic control as a few studies have found variation in the composition of lipids associated with chromatin isolated from different cells. For example, compared to the active chromatin from other cell types, the silent chromatin from sperm or avian erythrocytes contained lower amounts of tightly bound neutral lipids (e.g. cholesterol) and some PLs (e.g. CL), but it was mainly composed of PE [45,46]. Lipid components of chromatin in rat liver were reported to be dependent on the cell cycle [47].

Interesting possibilities arise from recent discoveries suggesting that lipids forming distinct microdomains can interact with subnuclear structures [48]. Electron microscopy analysis showed that SM is present in nuclear domains active in DNA replication, transcription, and possibly in different steps of mRNA processing as well [49]. On this basis, it was proposed that nuclear lipid microdomains might serve as a platform for transcription. These were suggested to be the anchoring point of the active chromatin in liver cells [50]. In a recent study of normal and cancer cells from liver, the authors found differences not only in the lipid composition of the nuclear lipid rafts and lipid microdomains, but also in the proteins involved in carcinogenesis associated with these structures, such as STAT3, Raf1, and PKCζ [51].
Interesting additional possibilities are offered by covalent lipid modifications of proteins or lipidation, which will be further reviewed.
LIPIDS IN DIRECT ASSOCIATION WITH DNA AND CHROMATIN
As early as the 1950s, a lipid was observed, in complex with nucleohistones prepared from calf thymus [52]. In the same report, the authors state that the same lipid was sometimes co-purified with isolated histones, but not with the DNA alone, suggesting that that lipid could be bound to histones. SM was proposed as a possible candidate based on chemical evidence from previous studies [53,54].
A possible function for such modifications may be suggested by a few studies showing that acidic PLs (including CL, PS, PI, and PG) promote more open transcriptionally permissive chromatin structures. In contrast, neutral lipids (PC, PE, and SM) appear to act in the opposite way, condensing chromatin fibers [19,55]. Thus, lipid modification of histones could contribute similarly to epigenetic marks such as histone acetylation and methylation.
Nonetheless, it also appears that lipids can directly bind DNA as well. In vitro studies with synthetic DNA polynucleotides reveal that neutral lipids (oleic acid and cholesterol) can be bound tightly to the DNA double helix [56] but not to triplex DNA. Spectroscopic results indicate that oleic acid shows molecular recognition of AT motifs by minor groove binding. In silico modeling suggests that PL binding to the minor groove is energetically favorable, with input from hydrogen, van der Waals, and hydrophobic bonds [56–58]. Since there is a lipid fraction in the nucleus, apparently tightly bound to DNA, the possibility of a new informational level in genomic DNA (“lipid code”) was postulated: lipids might specifically bind to the DNA in a sequence-specific way, and have significant functional roles for genome regulation. We will next discuss ways to understand this information, and what are the putative functions of the genome regulation via lipids.
LIPID MODIFICATIONS OF CHROMATIN PROTEINS
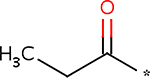
Covalent post-translational modifications greatly expand the functional protein repertoire of cells, allowing dynamic regulation of protein function, level of activity, localization, binding partners, and many more [59]. Acylation of lysine residues represents a notable class of modifications. The most famous modification of this class, acetylation, was discovered on histone tails. Acetylation would change histones’ charge and consequently their binding partners [60]. Since then, it has become clear that most proteins in eukaryotes are acetylated [61]. Lysine propionylation and butyrylation were also found on yeast histones less than a decade ago [62,63], and their addition to lysines was predicted to promote significant hydrophobicity to the proteins with important implications for their structure and function (see Figure 2). Furthermore, lysine malonylation and succination were discovered very recently by the combination of mass spectrometry and bioinformatics [64]. Succinyl-CoA and malonyl-CoA are important metabolites in the tricarboxylic acid cycle and fatty acid biosynthesis, and therefore the effect of their coupling to chromatin might be twofold: changing the properties of chromatin and removal of these moieties from the metabolic pool [65].
It has proven to be problematic to identify long-chain fatty acids as covalent attachments on proteins, presumably due to methodological complications such as the use of polar solvents in chromatin isolation methods, which prevents the enrichment of hydrophobic compounds like lipids. However, studies by Zou et al. [66] and Wilson et al. [67] provided two different approaches in identifying two long fatty acylations on histones H4 and H3.
In the former study, lysophosphatidylcholine acyltransferase-I was shown to translocate to the nucleus in the presence of calcium ions, to mediate O-palmitoylation of serine 47 on histone H4. Surprisingly, this modification was found to increase global levels of activated PolII, and therefore positively regulate transcription [66]. In the latter study, chemical reporters were used to detect a range of fatty acids (myristic, palmitic, and stearic) covalently coupled to a large number of nuclear and cytosolic proteins [67]. Particularly interesting was the finding that histone H3 variants were S-palmitoylated on cysteine 110, which could affect the ability of H3 to form a tetramer with H4, which in turn would have radical consequences for the affected chromatin. The attractive possibility is that this modification might change the binding properties of the modified chromatin to the nuclear membrane and nuclear lamins, as their carboxyl-terminal tails interact with H3 [68]. Lipid modifications of histones are summarized in Table 2.
Regulation of chromatin function by protein lipidation is not likely to be limited to histones. Palmitoylation of the heterochromatin-associated RAP1-associated protein Rif1 in yeast leads to its relocalization to the nuclear periphery, which increased silencing of the associated peripheral chromatin loci [69]. However, a more recent study indicated that palmitoylation is nonessential for this function of Rif1 [70], suggesting that the nuclear envelope relocalization is sufficiently important that multiple mechanisms have been evolved to target Rif1 to the periphery.
Finally, there are other lipid modifications of proteins, such as geranylgeranylation, phosphatidylethanolamination, and glycosylphosphatidylation [71]. With the wide range of lipid modifications already found on chromatin, it would not be surprising if these remaining protein lipid modifications could also be observed on chromatin, when they are studied in the future.
We expect that in the near future, using the combination of proteomic and lipidomic methods of chromatin purification will allow its isolation in the form of the intact “liponucleoprotein” to allow complete classification of lipids and fatty acids covalently linked to the amino acids. Even more exciting will be the understanding of the functions of these modifications, similarly to incredible variety of simpler moieties such as methylation, which was long considered to be inert (Prof Colyn Crane-Robinson, personal communication).
TOWARD UNCOVERING THE ROLES LIPIDS PLAY IN EPIGENETICS: FROM METABOLIC REGULATION OF GENE EXPRESSION TO A METABOLOEPIGENETIC CODE?
The natural fatty acids butyrate and lactate affect chromatin structure and function leading to the inhibition of histone deacetylase enzymes (HDACs), as well as DNA methylation, thus resulting in global changes to gene expression and chromosome functions [72,73]. In a similar context, there have also been reports suggesting a role for HDACs in lipid metabolism. However, lipid and fatty acid biosynthesis seems to be connected with epigenetic regulation more broadly.
Levels of acetyl-CoA are critical for the proliferation of eukaryotic cells [74], as it is a source of ATP in the carbon cycle, as well as the source of carbon for the production of lipids, fatty acids, and sterols. Moreover, it is necessary for histone acetylation in yeast [75] and for tumorigenesis [76].
Co-dependance of both lipid synthesis and histone acetylation on the same pool of acetyl-CoA is illustrated by a few lines of evidence. First, as with most genes, there is regulation of transcription of acetyl-CoA carboxylase, and other metabolic genes by histone acetylation/deacetylation in the promoter regions of these genes [77]. Second, direct protein acetylation of the metabolic proteins, including ones responsible for lipid biosynthesis, is important for their activity [61]. Third, it was recently shown that acetyl-CoA synthetase (ACSS2) is overexpressed in a large proportion of human tumors and is responsible for both lipid synthesis and acetylation of histones [76].
Like acetyl-CoA, 1-carbon metabolism is also crucial for lipid synthesis [78]. S-adenosine methionine (SAM) is a common donor of the methyl group for the processes of methylation of DNA (DNMT1 and DNMT3 in mammals), proteins (such as MLL3, Suv39, and Ezh2), and lipids (e.g. PE N-methyltransferase [79]).
The very exciting finding that methionine adenosyltransferase (MAT) is recruited to specific genes to provide SAM locally for usage by the repressive methyltransferases [80] suggests that at least in some cases chromatin modifications can happen using locally produced substrates to avoid competition with the cytosolic pool. This idea resonates with the aforementioned extensive presence of lipid metabolism and biogenesis enzymes within the nuclear membrane. In addition to the maintenance of the membrane itself, this could serve to provide the repertoire of chemical moieties required for the modification of nuclear proteins, including chromatin proteins. It will be intriguing to determine which metabolites contribute to the acetylation and methylation of proteins including histones, as it will have important implications for cancer epigenetics and the targeting of these marks in pharmacological therapies. Further experiments in the emerging field of metaboloepigenetics are warranted based on recent findings, and should be possible due to recent developments in metabolomics, proteomics, and transcriptomics.
Another dimension for lipid control over gene regulation has recently opened up due to the identification of the covalent “lipidation” of chromatin proteins discussed earlier. An attractive possibility is that locus-specific lipidation of chromatin allows hydrophobic interactions with specific regions of the inner nuclear membrane and nuclear lipid rafts, and therefore functional control through nuclear compartmentalization and packaging together with other lipidated chromatin domains, and/or temporal changes in chromosome territories. If the direct modification of DNA by lipids is subsequently shown in eukaryotes, a similar regulation by DNA–membrane interactions would be possible.
While we still need more extensive data on direct interaction between DNA and chromatin with specific lipids to properly model the roles of these interactions, it is an attractive hypothesis that, as the nuclear lipids surround chromatin, they affect its condensation, mobility, and accessibility, making it more or less transcriptionally permissive, thus epigenetically regulating gene expression and other processes involving chromatin. Since the concept of epigenetics and the histone code was introduced [81], many more histone modifications have been found, and it is currently not clear where chromatin modifications are the real regulators of the chromosome transactions and where they are simply signposts indicating an ongoing process due to other factors they recruit. Although adding lipid modifications to this already extremely complex gene regulation picture further increases its complexity, it may also be the only way we can hope to eventually understand how cell fates are chosen in normal development and how they are disrupted in many human diseases.