INTRODUCTION
Prion disease is a type of transmissible spongiform encephalopathy (TSE) that affects a species of mammals, such as, Creutzfeldt-Jacob disease (CJD) in humans, scrapie in sheep and goats, bovine spongiform encephalopathy (BSE) in cattle, and chronic wasting disease (CWD) in deer [1, 2]. The etiologic agent for prion disease is the prion, an abnormal conformational scrapie-like prion protein (PrPSc) converted from a normal host cellular form of prion protein (PrPC) in the central nervous system (CNS) [3]. The susceptibility, incubation period, and cross-species transmissibility of scrapie are mainly determined by the host prion protein gene (PRNP) [4, 5].
The Qinghai-Tibet Plateau is the highest plateau in the world, with an average altitude of 4000 meters. The Qinghai-Tibet Plateau is also the largest plateau in China (approximately 250 million km2). Because the animals of the Qinghai-Tibet Plateau are under the national first-class state protection, the Tibetan antelope (Rhinopithecus) is the only existing species of Tibetan antelope in the subfamily Bovidae that inhabits the alpine grasslands and desert areas at an altitude of 3250-5500 meters and is mainly distributed in the Qinghai-Tibet Plateau, which is centered in Qiangtang, China. Blue sheep (Pseudois nayauris) is the only species of blue sheep in the Bovidae subfamily, which can be divided into sichuan and xizang subspecies. Blue sheep are national second-class protected animals, are mainly distributed in the Qinghai-Tibet Plateau and the adjacent mountain areas at an altitude of 3200-5000 meters, and the habitat environment is mostly in areas of alpine bare rock and cliffs. Plateau pika (Ochotona curzoniae) is a small non-hibernating herbivorous mammal that is also known as black-lipped pika and belongs to the family Pika in the order Rabiformes. Plateau pika mainly live in the areas of alpine meadows and grasslands at an altitude of 3100-5100 meters. There are no reports of naturally-occurring TSEs in Tibetan antelope, blue sheep, and plateau pika. Furthermore, the PRNP genes have not been described in the literature.
In the present study, for the first time we obtained and sequenced the PRNP genes from the liver tissues of 21 Tibetan antelopes, 4 blue sheep, and 3 plateau pikas that had natural deaths in the natural reserve region of the Qinghai-Tibet Plateau. The PRNP genes of the Tibetan antelope and blue sheep were 771 bp long and encoded 256 aa (aa), and had 100% homology the with wild-type sheep prion protein (PrP) aa sequence. The PRNP gene of the pika was 759 bp long and encoded 252 aa with relatively high homology (92.1%) in the aa sequence of rabbits. Furthermore, the full-length prokaryotic recombinant PrP proteins of the sheep (rSheepPrP25-234) and pika (rPikaPrP23-230) were expressed and purified. Using the different dilutions of the brain homogenates of scrapie 263K-infected hamsters, scrapie 139A-, ME7- and S15-infected mice as the seeds, rSheepPrP25-234 and rPikaPrP23-230 were evaluated using real-time quaking-induced conversion (RT-QuIC) assays and controlled by the full-length hamster PrP (rHaPrP23-231). Positive reactions were noted in the preparations of rSheepPrP25-234 and rPikaPrP23-230, which showed different reactogenicity. Protein misfolding cyclic amplification (PMCA) tests of the brain homogenates of domestic sheep and rabbits with the seeds of scrapie strains 263K and ME7 failed to produce proteinase K (PK)-resistant PrP (PrPres).
MATERIALS AND METHODS
Ethics approval
The present study was approved by the Ethics Committee of the National Institute for Viral Disease Control and Prevention (Beijing, China) under protocol number 2009ZX10004-101, including the use of stored brain samples of scrapie agent-infected rodents and liver tissue samples of Tibetan antelopes, blue sheep. and plateau pikas.
Polymerase chain reaction (PCR) amplification and sequencing of the PRNP gene
The liver tissues of 3 Ochotona curzoniae (pika), 21 Tibetan antelopes, and 4 blue sheep that had natural deaths were collected from the Qinghai-Tibet Plateau in Qinghai province, China. Total DNA was individually extracted with a QIAamp DNA Mini Kit (QIAGEN, Limited Liability Company, Shanghai, China). The full-length PRNP sequence for each sample was amplified using the PCR technique with designed primers, including the primers for pika (upstream: 5′-ATGGCACACCTCAGCTACTGG-3′; downstream: 5′-TCATCCCACTATCAGGAAAA-3′) and antelope and blue sheep (upstream: 5′-GCCACTGCTATACAGTCATTCA-3′; downstream: 5′-ACTACAGGGCTGCAGGTAGA-3′). The reaction conditions were 94°C for 1 min, 52°C for 30 s, 72°C for 40 s for a total of 35 cycles. The PCR products were subjected to sequencing after purification according to the protocol described previously [6].
DNA and aa sequence alignment
The PRNP sequencing data and corresponding PrP aa sequences for each tested sample were collated and analyzed with software (DNAMAN 9.0; company, city, state, country). Pika, Tibetan antelope, and blue sheep PRNP nucleotide and aa sequences were compared with the PRNP nucleotide and aa sequences of other mammals using the Clustal W method and DNAStar 7.0 software (company, city, state, country). The homology matrix and phylogenetic tree based on a sequence alignment were further constructed.
Construction of recombinant plasmids
Recombinant plasmids expressing the full-length hamster PrP (rHaPrP23-231) were described previously [7]. To generate the recombinant plasmids expressing the full-length pika (rPikaPrP23-230) and sheep PrP (rSheepPrP25-234), the PRNP sequence of sheep was obtained by PCR with the following primers: forward (5′-TTCCATATGAAGAAGCGACCAAAACCTGGC-3′ with the Nde-1 site); and reverse (5′-CGGAATTCACTTGCCCCCCTTTGGTAAT-3′ with the EcoRI site). The pika PRNP sequence was obtained with following primers: forward (5′-TTCCATATGAAGAAGCGGCCAAAACCCGGAG-3′ with the Nde-1 site); and reverse (5′-CGGAATTCACTGGCCGCCCTCTGGT-3′ with the EcoRI site). The PCR products were cloned into a T-vector. After sequence verification, the PRNP fragments were released from the cloning vectors with Nde-1/EcoRl digestion and inserted into a prokaryotic-expressing vector (pRSETA; catalog no. V35120; Thermo Fisher Scientific, Inc., Waltham, MA, USA), generating p-SheepPrP25-234 and p- PikaPrP23-230 plasmids.
Protein expression and purification
Recombinant plasmids expressing rPikaPrP23-230, rSheep25-234, and rHaPrP23-231 were separately transformed into BL21(DE3) pLysS competent cells (catalog no. C1500; Beijing Solarbio Science & Technology Co., Ltd., Beijing, China). After ultrasonic treatment of the bacteria pellets, the lysates were denatured with guanidine-HCl and the target proteins were purified by Ni-NTA Superflow resin chromatography (catalog no. 30430; Qiagen, Hilden, Germany). The purified proteins were dialyzed into 10 mM sodium phosphate buffer (pH 5.8) and the concentrations of PrP proteins were adjusted to 500 μg/ml, as determined by absorbance measured at 280 nm. Following filtration (0.22 μm syringe filter, catalog no. SLGP033RB; Merck Millipore, city, state, country), the recombinant PrP proteins were aliquoted and stored at −80°C.
Brain samples of scrapie-infected experimental rodents
Brain samples from hamsters inoculated intracerebrally with scrapie agent 263K and brain samples from C57BL/6 (C57) mice inoculated intracerebrally with scrapie strains 139A, ME7, and S15 were enrolled in this study. The bioassay procedures, and the clinical, neuropathologic, and pathogenic features of the infected animals were described previously [8, 9]. The average incubation periods of 263K-infected hamsters and 139A-, ME7-, and S15-infected mice were 80.1±5.7 days, 183.9±23.1 days, 184.2±11.8 days, and 175.4±1.0 days, respectively. Age-matched healthy hamsters and mice were used as controls.
Preparation of brain homogenates
Brain homogenates were prepared according to the procedure described previously [9]. Brain tissues from the scrapie-infected and healthy rodents were thrice-washed in PBS and 10% (w/v) brain homogenates were prepared in cold lysis buffer (100 mM NaCl, 10mM EDTA, 0.5% Nonidet P-40, 0.5% sodium deoxycholate, and 10 mM Tris [pH 7.5]) containing a mixture of protease inhibitors (539134; Merck, city, state, USA). The tissue debris was removed with low-speed centrifugation at 2000 g for 10 min and the supernatants were collected for further study.
RT-QuIC assays
The details of the RT-QuIC assay were described previously [10]. Briefly, RT-QuIC reactions contained 1 μl of 10−5-, 10−7-, and 10−9-diluted brain homogenates from scrapie agent-infected rodents (1× PBS, 170 mM NaCl, 1 mM EDTA, 0.01 mM ThT, 0.001% SDS, and 10 μg of rSheepPrP25-234, rPika23-230, or rHaPrP23-231 [concentration: 1.03, 1.17, and 0.6 mg/ml, respectively] in a final volume of 100 μl). Each sample was assayed in quadruplicate. The assay was conducted in a black 96-well, optical-bottomed plate (265301; Nunc, city, state, country) on a BMG FLUOstar plate reader (BMG Labtech, city, state, country). The main working conditions were fixed, as follows: temperature, 50°C; vibration speed, 900 rpm; vibration/incubation time, 90/30 s; and total reaction time, 90 h. ThT fluorescence (excitation wavelength, 450 nm; emission wavelength, 480 nm) for each reaction was automatically measured every 30 min and expressed as relative fluorescence units (rfus). The cut-off value was set as the mean value of the negative controls plus 3 times the standard deviation. Each assay was repeated at least three times and Spearman-Karber analysis was used to estimate a seeding dose (SD50). The SD50 was calculated using the following formula:
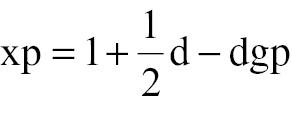
where xp = 1 represents the highest log dilution giving all positive responses, d is the log dilution factor, p is the proportion positive at a given dose, and gp is the sum of values of p for xp = 1 and all higher dilutions.
PMCA
PMCA was performed in a sonicator (Misonix sonicator 3000 and 4000; Misonix, Farmingdale, NY, USA) containing an adjustable temperature water bath circulation system and a microplate horn for PCR tubes. The hamster-derived prion seed (from 263K-infected hamster brains) and mouse-derived prion seed (from ME7-infected mouse brains) were diluted with the individual substrate of 10% brain homogenates prepared from healthy hamsters or mice, respectively. Ten microliters of different concentrations of seeds were mixed with 90 μl of substrate in thin-walled 0.2-ml PCR tubes containing 100 μl and placed in a floating position in the sonicator. One PMCA cycle included a sonication process at PM 9.5 for 20 s and an incubation process at 37°C for 29 min and 40 s. In this study, a complete direct PMCA was 80 cycles for mouse-derived prion seeds.
Western blot analysis
Brain homogenates and PMCA products were separated on 12% SDS-PAGE and electronically-transferred to nitrocellulose membranes with a semi-dry unit. After blocking with 5% non-fat dried milk in Tris-buffered saline (TBS) at 37°C for 1 h, the membranes were incubated at 4°C overnight with a PrP-specific monoclonal antibody [mAb] (SAF32, 1:5000 dilution; Cayman Chemical, Ann Arbor, MI, USA) at 37°C for 1 h. After washing with TBS-containing 0.1 % Tween-20 (TBST [pH7.6]) 4 times, the membranes were incubated with horseradish peroxidase-conjugated anti-mouse antibody (1:5000 dilution; company, city, state, country). Immunoreactive signals were developed using an enhanced ECL kit (PE Applied Biosystems, Foster City, CA, USA).
RESULTS
PrP sequences of the Tibetan antelope and blue sheep
Positive PCR products were obtained from 21 tested Tibetan antelopes and 4 blue sheep. Sequencing assays showed that the PRNP genes were 771 bp long, which may encode peptides 256 aa in length (submitted to GenBank, MW194318 and MW194319, respectively). The PRNP sequences of 21 tested Tibetan antelopes were highly homologous, with only one nucleotide difference (C297T, 10:11) that was a synonymous substitution. The 4 tested blue sheep also had highly homologous PRNP sequences, with one synonymous substitution (A567G, 3:1).
The PRNP sequence homology of Tibetan antelope, blue sheep, and other mammals were analyzed and are summarized in Fig 1A. Both Tibetan antelope (99.5% or 99.4%) and blue sheep (100% or 99.9%) had the highest homologous PRNP sequence with sheep. Higher PRNP sequence homologies of Tibetan antelope and blue sheep were also identified with goats, Thomson’s gazelle, cattle, and red deer (>90%). Less PRNP sequence homologies were demonstrated with camels, dogs, humans, ferrets, and cats, while least PRNP sequence homologies occurred with hamsters, rabbits, and mice.

Homology matrices of the PRNP gene sequences and PrP amino acid sequences of various species of mammals and humans. (A) DNA sequences. (B) Amino acid sequences.
The PrP aa sequence homologies among different mammals are shown in Fig 1B. Tibetan antelope and blue sheep had 100% identical PrP aa sequences with sheep, >95% homology with goats (99.2%), Thomson’s gazelle (99.2%), deer (98%), and cattle (97.7%), >90% homology with camels (94.5%), dogs (94.1%), ferrets (93.8%), rabbits (92.9%), cats (91.4%), humans (91.3%), and pikas (90.5%), and <90% homology with hamsters (89.3%) and mice (89.2%).
PrP sequences of Ochotona curzoniae (pika)
Sequencing assays of the PCR products from the samples of 3 pikas showed the same PRNP sequence (759 bp long encoding 252 aa [submitted to GenBank, MT371369.1]). As shown in Fig 1A, pikas had the highest PRNP gene homology with rabbits (90.4%). The PRNP gene homologies of the other animal species and humans were < 90%. Analysis of the pika PrP aa sequence homology with other species revealed the highest homology with rabbits (92.1%). Relatively higher homologies (>90%) were demonstrated in Thomson’s gazelle (91.3%), Tibetan antelope (90.5%), blue sheep (90.5%), sheep (90.5%), camels (90.4%), red deer (90.1%), and cattle (90.1%; Fig 1B).
PrP aa variations in Tibetan antelope, blue sheep, and pika compared to other mammals
The exact PrP aa sequences from Tibetan antelope, blue sheep, and pika, as well as 13 species of mammals, are shown in Fig 2A. Variations in aa among various species mammals appeared in the signal peptide, α-helix 3, and GPI anchor regions. There were 23 and 30 aa differences between sheep and humans, and between pika and humans, respectively. Furthermore, a phylogeny tree was constructed to illustrate the distances in PrP a. sequences between various species (Fig 2B). Tibetan antelope and blue sheep shared 100% homology in PrP aa sequences with sheep, and showed high homology with goat and other artiodactyls (Thomson’s gazelle, deer, and cattle), while an apparently remote relationship among the animal species with naturally-occurring prion disease (cats, camels, and humans) and routinely-used experimental animals (hamsters, mice, and ferrets), as well as prion-resistant animals (dogs and rabbits). Pika revealed a relatively remote PrP aa sequence relationship with other species, including rabbits.
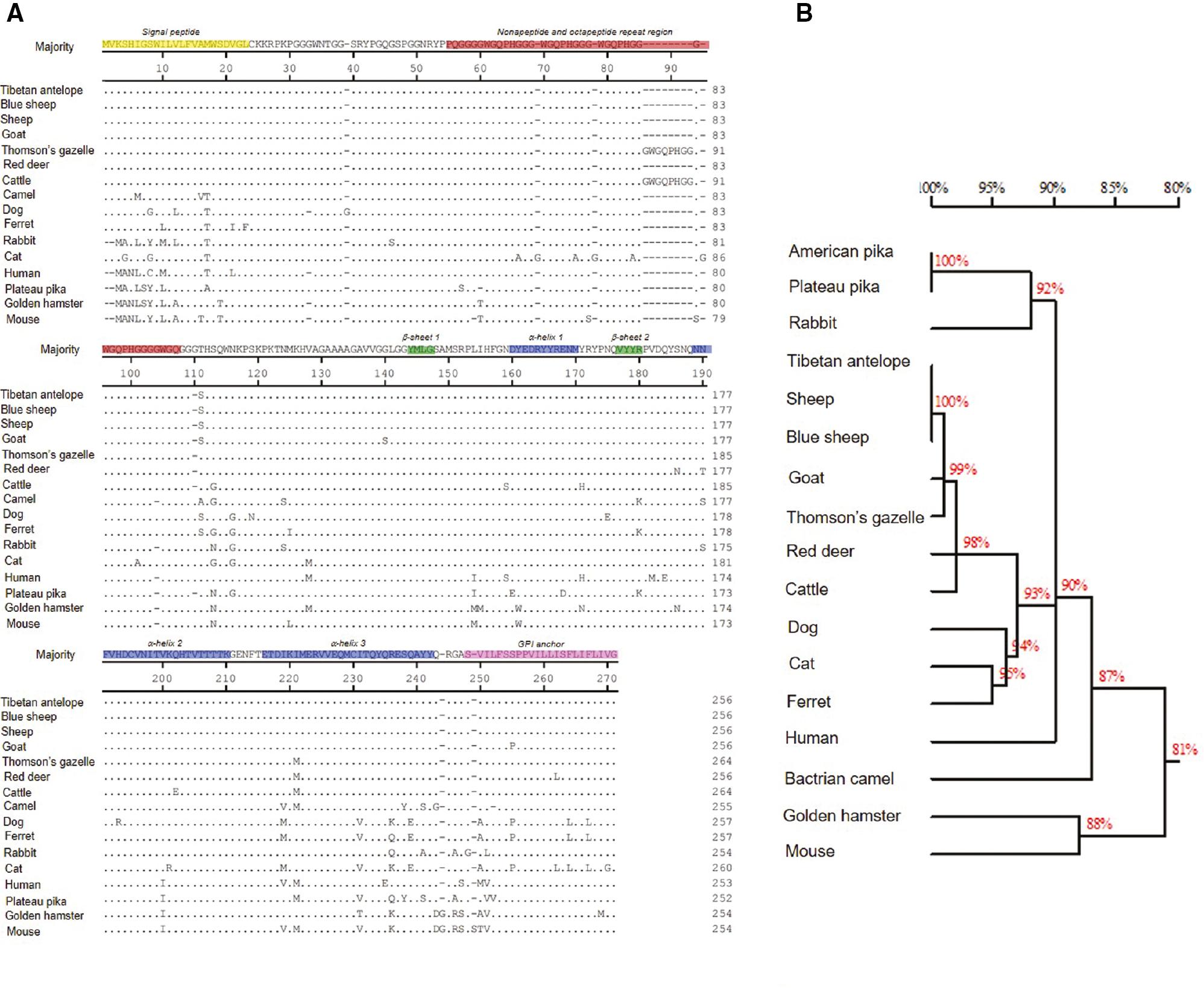
Comparison of the variations of PrP amino acids of various mammals and humans. (A) Full-length amino acid sequences of PrPs. The various functional and secondary structural regions within PrP sequences are indicated with colors. The amino acid numbers are shown on the right. (B) Phylogenetic tree based on amino acid sequences of PrPs.
Hamster and mouse-adapted scrapie agents efficiently induced conversion of the recombinant blue sheep and pika PrPs on RT-QuIC
To test the feasibility and reactivity of the PrPs from Tibetan antelope, blue sheep, and pika in the Qinghai-Tibet Plateau on RT-QuIC, full-length recombinant PrPs based on the PRNP sequences of blue sheep and pika were expressed and purified from E. coli. Because the PrP aa sequences of Tibetan antelope and blue sheep shared 100% homology, we only prepared the prokaryotic recombinant PrP blue sheep. SDS-PAGE and PrP-specific Western blot analysis identified a single band at approximately the 21 kDa position in the rSheepPrP25-234 and rPikaPrP23-230 preparations (S1 Fig).
The same amounts of rSheepPrP25-234, rPikaPrP23-230, and rHaPrP23-231 were mixed with different prion strains at different dilutions (10−5, 10−7, and 10−9), then subjected to RT-QuIC assays. Because rHaPrP23-231 was the routinely used substrate in our RT-QuIC assays, which revealed reliable reactivities in the reactions using the brain homogenates of hamsters and mice infected with various scrapie strains as the seeds at dilutions >10−7, only the rHaPrP23-231 reactions with 10−7 and 10−9 diluted scrapie strains were performed. A 10−5 diluted brain homogenate of normal hamster was used as a negative control. In the preparation of 10−5 diluted scrapie strains, both rSheepPrP25-234 and rPikaPrP23-230 revealed positive reactive curves with all four tested scrapie strains (263K, 139A, ME7, and S15 [Fig 3A–D, respectively]). In the preparation of 10−7 diluted scrapie strains, 4 wells of rSheepPrP25-234 had positive curves with 263K, while only 1 of 4 wells displayed a positive reaction after a long lag with 139A and S15 and all 4 wells were negative with ME7. In contrast, positive reactive curves were observed in all four rPikaPrP23-230 wells with 263K, 139A, and S15, and in 3 of 4 wells with ME7. Similarly, positive curves were noted in all rHaPrP23-231 reactions with all four scrapie strains. In the preparation of 10−9 diluted scrapie strains, only one rHaPrP23-231 well revealed a positive curve with 139A, and the remaining reactions were all negative. The SD50 values of strains 263K, 139A, ME7, and S15 in RT-QuIC using rSheepPrP25-234 as a substrate were 1010.25±0.5/g, 1010.5±0.75/g, 1010±0.5/g, and 1011.125±0.375/g, respectively, while the SD50 values in RT-QuIC using rPikaPrP23-230 as a substrate were 1016±0.75/g, 1012.25±0.5/g, 1012.63±0.13/g, and 1012.38±0.38/g, respectively (Table 1).
Average SD50 values of three species recombinant PrPs with 4 scrapie strains.
| PrP protein | SD50 | ||||
|---|---|---|---|---|---|
| Strain | 263K | 139A | ME7 | S15 | |
| rSheepPrP25-234 | 1010.25±0.5/g | 1010.5±0.75/g | 1010±0.5/g | 1011.125±0.375/g | |
| rPikaPrP23-230 | 1016±0.75/g | 1012.25±0.5/g | 1011.5±0.5/g | 1012.375±0.375/g | |
| rHaPrP23-231 | 1012.63±0.13/g | 1012.25±0.25/g | 1012.625±0.125/g | 1012.625±0.126/g | |

RT-QuIC assays of rSheepPrP25-234, rpikaPrP23-230, and rHaPrP23-231 to four different rodent-adapted scrapie strains. (A) Hamster-adapted strain 263K. (B) Mouse-adapted strain 139A. (C) Mouse-adapted strain ME7. (D) Mouse-adapted strain S15. The dilutions of scrapie strains are shown in the top of each graph. Neg Ctrl: negative control of 10-5 diluted brain homogenate of normal hamster. Blank Ctrl: blank control of PBS. Various recombinant PrP proteins are indicated on the right.
The average lag times and the rfu peaks of the three species recombinant PrPs with four different scrapie strains at dilutions of 10−5 and 10−7 are summarized in Table 2. Both rSheepPrP25-234 and rpikaPrP23-230 were positive in 10-5 dilutions of all strains and reached the rfu detection limit (260,000) in our RT-QuIC. The lag times of rSheepPrP25-234 were clearly longer than rpikaPrP23-230. In contrast to rSheepPrP25-234, RT-QuIC was positive at a 263K 10−7 dilution with a relatively long lag phase (24.3 h). Both rpikaPrP23-230 and rHaPrP23-231 elicited positive reactions in the presence of the four scrapie agent 10-7 dilutions. Moreover, rpika23-230 had notably shorter lag times and higher rfu rHaPrP23-231 peaks in the reactions with all tested four scrapie strains.
Average lag times and rfu peaks of three species recombinant PrPs with 4 scrapie strains at 10−5 and 10−7 dilutions.
| PrP protein | Dilution | 10−5
| 10−7
| ||||||
|---|---|---|---|---|---|---|---|---|---|
| Strain | 263K | 139A | ME7 | S15 | 263K | 139A | ME7 | S15 | |
| rSheepPrP25-234 | Average lag time (h) | 13.8±0.9 | 14.2±4.3 | 21.9±16.8 | 10.9±1.9 | 23.6±12.8 | - | - | - |
| rfu peak (X1000) | 260 | 260 | 260 | 260 | 260 | - | - | - | |
| rPikaPrP23-230 | Average lag time (h) | 4.4±0.2 | 5.2±2.5 | 4.3±0.3 | 3.8±0.6 | 12.9±3.8 | 6.0±1.4 | 13.3±6 | 18.6±12.4 |
| rfu peak (X1000) | 260 | 260 | 260 | 260 | 260 | 260 | 260 | 260 | |
| rHaPrP23-231 | Average lag time (h) | ND | ND | ND | ND | 19.3±8.7 | 8.6±1.7 | 17.5±8.8 | 23.7±18.5 |
| rfu peak (X1000) | ND | ND | ND | ND | 204±9.1 | 165±10.0 | 211±17.4 | 181±8.0 | |
ND: not done.
Hamster- and mouse-adapted scrapie agents failed to induce conversion of the native brain PrPC from sheep and rabbits based on PMCA
Because Tibetan antelope, blue sheep, and plateau pika are nationally-protected wild animals, it is impossible to obtain fresh brain tissues. To evaluate the possible conversion activities of mouse- and hamster-adapted scrapie agents on brain PrPC of plateau animals based on PMCA, the brain homogenates of domestic sheep and rabbits were prepared and used as mimics. As expected, clear PK-resistant PrP signals (PrPres) were noted in the reactions using scrapie strain 263K as the seed and hamster brain homogenate as the substrate (10−4; Fig 4A, upper panel) and using scrapie strain ME7 as the seed and mouse brain homogenate as the substrate (10−3; Fig 4B, upper panel). Under our experimental conditions, no PrPres signal was detected in any reaction involving scrapie strain 263K (Fig 4A, lower panel) or ME7 (Fig 4B, lower panel) mixed with brain homogenates of sheep (left) or rabbits (right). Furthermore, two rounds of blind passages of the PMCA products to the brain homogenates of sheep and rabbits in PMCA failed to identify the PrPres signal. This finding implies that the prions in the brain tissues of scrapie-infected experimental hamsters and mice are incapable of inducing conversion of brain PrPC in domestic sheep and rabbits based on PMCA.

PMCA tests of the brain homogenates of domestic sheep and rabbits with two rodent-adapted scrapie strains. (A) Hamster-adapted scrapie strain 263K. (B) Mouse-adapted strain 139A. Above panels are the PMCA tests with homologous brain substrate and seed. The PMCA products were subjected to PrP-specific Western blots with (+) or without (−) digestion of proteinase K (PK). BH: brain homogenate.
DISCUSSION
In the current study, for the first time we report the PRNP sequences of three wild mammals from the Qinghai-Tibet Plateau (Tibetan antelope [Rhinopithecus], blue sheep [Pseudois nayauris], and plateau pika [Ochotona curzoniae]). Tibetan antelope and blue sheep were shown to have extremely high homology in the PRNP gene and 100% identical homology in the PrP aa sequences with sheep. Two novel synonymous polymorphisms have been identified (C297T in Tibetan antelope and A567G in blue sheep) involving the encoding codon for G66 in the 1st octarepeat and N189 in the 2nd α-helix. No polymorphisms were found in the three pika samples. Because of the limited number of blue sheep and pikas in this study, the presence of other PRNP gene polymorphisms cannot be absolutely excluded.
It is well-known that the polymorphisms at codons 136, 154, and 171 in sheep PrP are closely associated with susceptibility to scrapie; specifically, VRQ is highly sensitive, ARQ is moderately sensitive, and ARR is less sensitive [11–14]. All tested Tibetan antelope and blue sheep samples in this study had the ARQ genotype at codons 136, 154, and 171. This finding highlights that those two species are potentially susceptible to scrapie. The Tibetan antelope and blue sheep ARQ genotype is based on a study with a limited number of tested individuals obtained from a limited geographic regions And cannot be extended to the entire population in the Qinghai-Tibet Plateau.
Plateau pika is a species endemic to the Qinghai-Tibet Plateau in large numbers. Plateau pika has very high homology with American pika (Ochotona princeps) for the PRNP gene, but a relatively remote association with other tested mammals, including rabbits with the highest homology (92.1%). The susceptibility of pika to scrapie or other prion agents is unknown and hard to predict. Plateau pika is considered to have an important role in maintaining biodiversity and ecosystem balance in the Qinghai-Tibet Plateau where there are approximately 210 wild mammal species. Numerous caves excavated by pikas also provide nests for many small birds and lizards. The pika caves nourish the plants and provide ideal conditions for the diversity of plant species in this region. Pika is also the main prey of most small- and medium-sized carnivores, and almost all grassland raptors. As one of the important links in the biological chain in the Qinghai-Tibet Plateau, the importance of pika for prion circulation if pikas are susceptible to prion infection.
Our data verify that the full-length recombinant PrP proteins of both sheep and pika can be efficiently converted to positivity in the RT-QuIC assays in the presences of hamster- and mouse-adapted scrapie strains, which indicates that the PrP proteins of blue sheep, Tibetan antelope, and plateau pika are able to form fibrous structures from hamster and mouse prions under RT-QuIC experimental conditions. It is known that the efficient transmission between species depends largely, but not entirely, on the host PrP protein homology between donor and recipient [3, 15, 16]. Such phenomenona may also appear in in vitro tests for misfolding proteins, such as RT-QuIC and PMCA [17, 18]; however, breakthrough of the species barrier is much easier under RT-QuIC and PMCA conditions [18]. The RT-QuIC results here verified again a breakthrough of the species barrier despite an apparent difference in PrP aa sequences of sheep and pika in comparison with the donors (hamster and mouse).
We also found a difference in the RT-QuIC reactivities to three mouse-adapted scrapie strains between sheep and pika PrP proteins. Pika recombinant PrP had a higher RT-QuIC reactivity than sheep PrP. Pika PrP displayed slightly stronger reactivity than hamster PrP under our experimental conditions. The sheep recombinant PrP did not have stronger RT-QuIC reactivity to the scrapie strains used in the current study because all prion strains were rodent-adapted scrapie strains. Pika and sheep PrPs showed similar percentages of homology compared with mice and hamsters. In addition to the signal peptide and GPI anchor regions that were removed in the full-length recombinant proteins, the most obvious diversity of aa sequences between those two species were located in the 3rd α-helix region; the precise underlying molecular mechanism needs further exploration. Nevertheless, the good and stable reactivity of pika PrP to various rodent-adapted scrapie strains in RT-QuIC deserve further analysis for feasibility as a substrate in RT-QuIC tests, including the assays for human prion disease.
Generally, PMCA products possess both biochemical characteristics and infectivity of prions, whereas RT-QuIC products are more of a reflection of prion fibrillation [19–21], which usually lack infectivity [22]. Overcoming natural species barriers of prion infection is observed in the experimental bioassays of PMCA product inoculation. Our previous study confirmed that inoculation of the newly-formed cross-species PMCA products of hamster-adapted (strain 263K) and mouse-adapted (strain 139A) scrapie agents were amplified with heterogeneous brain tissues (139A-hamster PMCA-PrPres and 263K-mouse PMCA-PrPres) to induce experimental TSEs in the other animal. In contrast, the homologous PMCA products (139A-mouse PMCA-PrPres and 263K-hamster PMCA-PrPres) showed no infectivity against the other animal [23]. Overcoming species barriers of prion infections by PMCA has been repeatedly documented to better understand the transmission between different species of animals and from animal-to-human [24]. Based on PMCA, PrPres can be generated in the reactions of different species of PrPSc with PrPC from rabbits and dogs that are considered as poorly susceptible and tolerant to prion infection [25, 26]. This finding strongly suggests that PMCA can facilitate prion strain breakthrough a species barrier and propagate in vitro and in vivo. Three blind passages of hamster and mouse prions in this study failed to induce conversion in the brain homogenates of domestic sheep and rabbits in PMCA. One direct technical possibility is the relatively limited amplifying rounds and times in PMCA. A further increase in PMCA rounds is warranted. In contrast, high efficiency of PMCA in circumventing species barriers lead to doubt that the species barriers in the real world are potentially underestimated; therefore, careful interpretation of the results of PMCA experiments are required [24]. Nevertheless, numerous species of ruminants have been verified to be susceptible to scrapie infection and bovine spongioform encephalopathy (BSE) [3, 27–29]. One may assume that Tibetan antelopes and blue sheep, showing high homology in PrP aa sequences with sheep, are also susceptible to natural scrapie infection.
In conclusion, this study is the first description of PRNP genes and PrP aa sequences of Tibetan antelope, blue sheep, and plateau pika on the Qinghai-Tibet Plateau. In the presences of rodent prions, the PrPs of sheep and pika efficiently induce fibrillation in RT-QuIC, but do not generate PrPres in PMCA. Our results indicate that pika, as one of the important links in the biological chain in the Qinghai-Tibet Plateau, may have an important role in prion circulation. Pika PrP deserves further analysis for potential application value in assays for human prion disease.
