Introduction
The present-day methodology of chromosomal dosimetry provides dose estimates which are sufficient in many radiological situations.[1] However, this methodology also contains limitations which lead to serious difficulties in cytogenetic data interpretation and large uncertainties in dose estimates for radiation exposure high enough to cause deleterious health effects or are life-threatening.[2] The worst outcomes of inaccurate dosimetry are underestimates of dose, jeopardizing appropriate clinical management, and overestimates of doses or false positive reports, leading to unwarranted anxiety and unnecessarily burdening the health infrastructure. Major causes of these problems include incorrect implementation of the cytogenetic procedures or lack of experimentally justified, clear instructions.
The crucial factor for the emergency biodosimetry is the speed of obtaining the dose estimates. However, the triage of victims can be performed successfully only if the aberration scoring has been done with sufficient accuracy.[3] The optimization of uncertainties in dose estimates through defining the required numbers of metaphases scored and aberrations found has been effectively resolved for low-dose radiation exposures using an expertise mode (manual microscope scoring of 1000 cells per sample) of cytogenetic analysis[4] and to some extent for a wider dose range of homogeneous exposure.[5] However, with one exception,[3] no such attempt has been made for partial-body exposure scenarios.
There are a growing number of reports regarding triage biodosimetry of partial-body exposures performed in experiments with in vitro simulations.[6789101112] These studies focused mainly on the accuracy of dose estimates and their efficacy in classifying samples between triage categories. Much less information can be found in the literature regarding the speed of collecting the necessary number of metaphases or aberrations in scenarios of inhomogeneous exposure. Furthermore, many authors presented comparisons of the accuracy of manual and automated cytogenetic analysis, but only a few studies[813] showed actual data on time required for dicentric quantification in each mode. There has only been one mention of the problems of accuracy and speed of biodosimetry from the standpoint of the aberration yield present in the sample,[14] but still there is no clear understanding of the shape of this possible dependence in scenarios, including different doses to different exposed fractions of the body.
Thus, further improvement of the methodology of cytogenetic triage of victims with suspected inhomogeneous irradiation entails a measurement of time required for getting aberration yield estimates at a certain level of accuracy, and vice versa, evaluating the achievable numbers of metaphases scored and aberrations found during a fixed time of microscopy. Obviously, such a study has to include a variety of exposure scenarios and must be done in well-controlled experimental conditions, minimizing possible sources of variations in the aberration scoring process. A special exercise was performed with an attempt to satisfy these criteria, and the current report presents the findings of this research.
Materials and Methods
The experiment was performed using the technical facilities of the Cytogenetics and Biomarkers Group of the Centre for Radiation, Chemical and Environmental Hazards of the Public Health England, Harwell, Oxfordshire, United Kingdom (UK). Blood was irradiated at the irradiation facility of the Genome Stability Unit of the Medical Research Council, Harwell, UK. Peripheral blood was taken from a healthy male volunteer, aged 39 years, with no occupational or medical exposure to any cytotoxic substances or clastogens, except occasional dental and chest X-rays. The donor's participation was with written informed consent and according to the Institutional Ethics Protocol. The details of blood irradiation and lymphocyte culturing were identical to those used earlier in the cytogenetic experiment on setting up a calibration curve for high radiation doses,[15] in accordance with the requirements given in the International Atomic Energy Agency (IAEA) Manual on chromosomal dosimetry.[1]
In the present work, the donor's blood was exposed to 1.98, 3.96, 11.88, and 20.3 Gy acute 60Co γ-rays, accompanied by a sham-treated zero-dose control sample. After irradiation, blood samples irradiated to ~2, ~4, and ~12 Gy were mixed with unirradiated blood to simulate partial-body exposure scenarios so that the blood fraction exposed to each dose comprised 90%, 50%, or 10% of the sample. The unmixed blood samples represented the homogeneous (100%) exposures to ~2, ~4, ~12, and ~20 Gy and a zero-dose control.
Lymphocyte cultures were set up from mixed and unmixed blood according to the standard technique.[115] Cultures were maintained in the presence of bromodeoxyuridine at 37.5°C for 52 h for experimental points of ~2, ~4, and ~12 Gy, but for ~20 Gy, the culture time was increased to 68 h. Cells were fixed and metaphase preparations were stained by the fluorescence-plus-Giemsa (FPG) method.
The fixed time microscopy was carried out on coded slides by one cytogeneticist, using the same microscope (Carl Zeiss Axioskop 20, Germany), to avoid any observer bias and possible inter-operator variability. Chromosomal damage was recorded in diploid cells; the in vitro mitosis number was identified by the patterns of FPG staining. Aberrations were classified using published criteria.[1] All kinds of chromosome and chromatid-type breaks and exchanges were recorded; however, the data analysis in the present report includes unstable chromosome types only. These comprised dicentrics (Dic) and centric rings (CR), accompanied by a fragment, and excess acentric fragments. Polycentrics were converted into the equivalent number of Dic.
The microscopy also included recording the number of metaphases analyzed by the operator during a fixed period, usually 70–100 min. Metaphases of poor quality, in which the chromosomes were unsuitable for analysis, were recorded as “rejected cells” for data completeness. Up to 13 rounds of fixed time microscopy was carried out per experimental point depending on the number of metaphases on the slide.
For statistical analysis, the general methods of variation, correlation, and regression analyses were applied. The aberration yield was expressed per cell. The regressions were fit by the unweighted least squares method using the respective option of the Microsoft™ Excel software package.
Results
During the exercise on fixed time cytogenetic analysis for biological dosimetry, 115 rounds of microscopy with a total duration of 153.4 h were carried out; 16 665 cells were scored (8535 recorded, 8130 rejected); 1681 aberrant cells were identified, in which 2975 Dic and centric rings (Dic + CR) were found. The averaged results of the exercise for each experimental simulation of total- or partial-body irradiation are shown in Table 1. In the present report, attention was focused on two parameters which technically define the accuracy of cytogenetic dosimetry: The number of recorded metaphases and the number of aberrations found. For both parameters, the acquisition rate was calculated as a ratio of the actual value divided by the duration of microscopy round (cell per min and Dic + CR per·min).

The numbers of cells scored and aberrations found during the exercise for fixed time microscopy for chromosomal dosimetry of simulated homogeneous and partial-body irradiation in vitro to various doses of acute γ-rays
The overall speed of the microscopy varied considerably between experimental points; the rate of metaphase recording ranged from 6 to 12 cell per·h in the scenarios of 12 or 20 Gy, 100%, to 110–115 cell per·h for localized exposures of 2–12 Gy, 10%. The speed of the operator's work depended on (i) the overall density of metaphases on a slide, (ii) ratio of “scorable” and “unscorable” metaphases, and (iii) Dic + CR yield in a sample.
For the first factor (i), it was found that there is little influence on the speed when the density of metaphases on the slide is within the optimal range of about 2–7 in each field of view at low microscope magnification (×90–100). This density can be adjusted by concentrating or diluting fixed cell suspensions before slide making. At lower density, time is wasted in searching for metaphases, and at higher density, there is a risk of scoring the same cell twice. Sides with densities outside of the optimal range could be analyzed using an automated system equipped with a metaphase finder.
The impact of the two latter factors, (ii) and (iii), was explored statistically by regression analysis [Figures 1–5]. The overall quality of the preparations, designated here as a proportion of rejected metaphases within the total number of cells, appeared not to be a crucial factor for the cell scoring rate [Figure 1]. The correlation in this dataset occurred mainly due to three points with rejected fractions <0.2 and three points with rejected fractions >0.8. If these points are excluded from the analysis, the correlation drops down to r = −0.180.

The dependence of the cell recording rate on the proportion of rejected metaphases among total cells found during the exercise of fixed time microscopy for biological dosimetry. The solid line represents the linear regression shown on the plot

The dependence of the cell recording rate on the yield of aberrant cells (YAbCells) in the sample during the microscopy for biological dosimetry. Each dot represents the values estimated during one round of the fixed time microscopy. The solid line denotes the exponential regression of the cell recording rate on the YAbCells, which equation is given on the plot
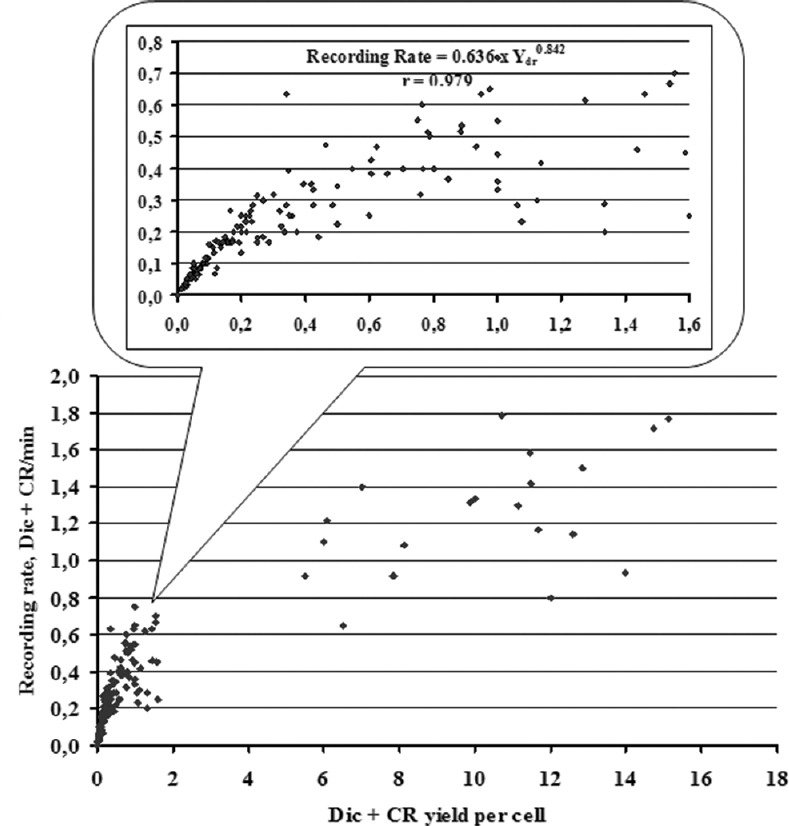
The dependence of dicentrics plus centric rings (Dic + CR) recording rate on Dic + CR yield (Ydr) in the sample during the fixed time microscopy for biological dosimetry. Each dot represents the values estimated during one round of fixed time microscopy. The pattern occurring at Ydr ranged from 0 to 1.6 per cell is shown in detail on the additional plot. The linear regressions were constructed separately for two distinctive intervals of Ydr: ≤1.6·per cell and >5·per cell; the latter corresponds to experimental points of homogeneous irradiations to 12 and 20 Gy. Within the latter cluster, the linear correlation was moderate: Dic + CR per min = 0.715 + 0.053 Ydr (r = 0.492), meanwhile in the main dataset, the linear fit was stronger: Dic + CR per min = 0.080 + 0.367 Ydr (r = 0,823). However, if zero values of Ydr are replaced by the mean background yield value of 0.8 × 10−3 per cell, then the dependence for Ydr ≤1.6·per cell can be effectively approximated by a power function, shown on the plot

The dependence of the cell recording rate on the yield of dicentrics plus centric rings (Dic + CR) in the sample during microscopy for biological dosimetry. Each dot represents the values estimated during one round of fixed time microscopy. The pattern occurring in the Dic + CR yield rang of 0–1.6 per cell is shown in detail on the additional plot. The exponential regressions shown on the plots were constructed separately for two distinctive intervals of Dic + CR yields: ≤1.6·per cell and >5·per cell; the latter corresponds to experimental points of homogeneous irradiations to 12 and 20 Gy. The solid line on the additional plot denotes the exponential regression of the cell recording rate of the yield of Dic + CR (Ydr)

The dependence of the cell recording rate on dicentrics plus centric rings (Dic + CR) recording rate during microscopy for biological dosimetry. Each dot represents the values estimated during one round of fixed time microscopy. The additional plot shows in detail the pattern formed by the main dataset excluding the experimental points of homogeneous irradiations to 12 and 20 Gy. The solid line and the exponential equation on the additional plot correspond to the truncated dataset. The dashed line and the equation on the main plot are of the exponential regression, generated on the full dataset
The overall relationship between the cell recording rate and the aberrant cell yield was negative, meanwhile that between Dic + CR acquiring rate and their yield (Ydr) appeared to be positive, both with a tendency to saturation at high yield values, observed on experimental points of ~12 Gy, 100% and ~20 Gy, 100% [Figures 2 and 3].
As a result, a strong dependence occurred for the cell scoring rate on the Ydr, measured during one round of microscopy [Figure 4]. This relationship was biphasic, with a quick exponential decline in the range of Ydr between 0 and 1.6 Dic + CR·per cell, followed by a slow decline in the interval of very high Dic + CR yields.
The relationship between the rates of recording scorable metaphases and detecting Dic + CR fits best to the exponential model [Figure 5]. The presence of the data from the ~12 Gy, 100% and ~20 Gy, 100% points in the analysis did not cause changes of the regression coefficient; thus, the overall decline in the cell scoring rate was constant within the studied range of Dic + CR per·min values.
The data collected during the fixed time microscopy were used (i) for computation of the time needed for scoring a certain number of cells or identifying a certain amount of cytogenetic damage, which gives the dose estimates at the triage or expertise level of accuracy, and (ii) for assessment of the accuracy of the analysis from the standpoint of the numbers of cells scored and aberrations found during the fixed time of the microscopy work.
The mode (i), “time versus accuracy,” included modeling several modes of the biodosimetry service: triage analysis of 50 cells, expanded triage mode of 100 cells, conventional scoring of 500 cells, and expertise level of 1000 cells, or stopping when 100 Dic + CR are reached, as recommended.[1] The Dic + CR acquiring rates were averaged for each of 13 experimental points, presented in Table 1, and the mean time needed for recording a desired number of cells or Dic + CR was calculated. The results are shown in Table 2.

Time required for reaching a certain accuracy of chromosomal analysis for biological dosimetry
The mode (ii), “accuracy versus time,” was done by modeling the workload of 1 case per week and 1, 5, and 10 cases per day per operator. The time of productive microscopy was assumed to be 5 h per day for one operator. Thus, for 1 case per week, this implies 25 h, for 1 case per day –5 h, for 5 cases per day –1 h, for 10 cases per day –0.5 h of scoring time allowed per sample. The accuracy was evaluated as a ratio of the error of the number of Dic + CR found to the number of Dic + CR found; the former was calculated assuming Poisson statistics for simplicity, i.e., as a square root of the latter. Thus, the error = (√X/X)·100%, where X is the number of Dic + CR. Again, the Dic + CR acquiring rate was averaged for each of 13 experimental points, and from this, the mean number of cells scored and the Dic + CR found for each fixed period were estimated [Table 3]. The relative error of the Dic + CR yield measurement during the fixed time of scoring was calculated and plotted against the duration of the analysis for each studied scenario of radiation exposure [Figure 6].

The productivity of chromosomal analysis depending on time available for microscopy of one sample

The accuracy of dicentrics plus centric rings (Dic + CR) measurement, achievable at different scoring times of the sample in various scenarios of radiation exposure. The accuracy was evaluated as a relative error (%) on the number of Dic + CR identified during 0.5, 1, 5 or 25 h of the analysis; that was computed as error = (√X/X)·100%, where X is the number of aberrations. The exposed blood fractions were 10% (1:9, blue circles), 50% (1:1, rose triangles), 90% (9:1, red squares), and 100% (1:0, black diamonds), as indicated on the plot for each of the radiation doses (~2, ~4, ~12, and ~20 Gy)
Discussion
The experimental research, presented here, was specifically performed to assess the accuracy of the cytogenetic analysis achievable for certain microscopy analysis times, and vice versa, to evaluate the time required for scoring a certain number of cells or identifying a certain number of aberrations in complex radiation exposure scenarios. The necessity of such a study was identified due to the extensive preparations of the international biodosimetry community for radiological emergencies. Since triage has become the main strategy in the initial phase of the biodosimetry response to a large-scale event, possible mistakes and unrealistic expectations at this step may have implications on the accuracy of the dose estimates. Cytogenetic laboratories performing practical biodosimetry must have clear guidelines for planning the schedule of microscopy work from the standpoint of “time versus accuracy” and “accuracy versus time” for different exposure scenarios and different workloads for the laboratory. However, there are no quantitative data in the literature which can be used for developing such protocols. In published reports, the problem of the accuracy of dose estimates dominates over the question of the time needed/available for the analysis.[6789101112] Therefore, the current work attempts to fill in this gap in the methodology of biodosimetry. In the present experiment, effort was made to minimize the influence of possible confounding factors such as the blood donor, lymphocyte irradiation and culture conditions, slide making and staining technique, microscope, and operator. This allowed for the data analysis to be focused mainly on radiobiological and cellular characteristics.
It is well known that one of the complications in biodosimetry after high-dose exposures is the low mitotic index of cultured lymphocytes occurring due to radiation-induced mitotic delay and intensive interphase cell death. The problem is partially solved by setting up replicate cultures, which can be combined during cell fixing to get a sufficient number of metaphases on a slide. A low concentration of metaphases leads to time lost due to scanning of the slide. We found that the microscopy becomes most productive in the optimal metaphase density range of 2–7 in any field of view at low magnification. This avoids excessive scanning and often allows moving from cell to cell at high magnification, without having to change objectives. For a very high concentration of metaphases, omitting the objective change is not possible as there is a risk of scoring the same cell twice. The optimal metaphase density can be technically achieved by concentrating or diluting the fixed cell suspension. Meanwhile, the slides with too low or too high metaphase numbers can be preferably scanned on an automated microscopy system equipped with a metaphase finder and precise positioning devices.
The most striking finding was that the overall speed of the radiation cytogenetic analysis, and thus of biodosimetry, depends primarily on the aberration yield which in turn depends on radiation dose and the irradiated fraction of cells. Earlier, it was found that the estimated time for sample analysis was highly dependent on the dose that the sample received; samples exposed to higher doses required more time for analysis, and generally, damaged cells were more difficult to score due to a requirement to ensure spread completeness.[14] No other study specifically mentioned this problem of accuracy and speed of biodosimetry from the standpoint of the aberration yield in the sample; thus, our report seems to be the first presenting direct estimates on this issue. The numbers of recorded metaphases varied from about 2 cells·per min at low Dic + CR yields, induced by localized exposure (irradiated fraction 10%), to 1 cell in 20 min (just 6 cells·per h) at very high yields resulting from homogeneous exposures to doses as high as 20 Gy. A strong negative correlation occurred between the rate of Dic + CR acquisition and the rate of recording cells. In practical settings, this makes it impossible to simultaneously acquire a high number of aberrations in a large sample of cells. Therefore, biodosimetry experts must always make a choice as to which is preferable in each particular case.
The data obtained in the exercise with the fixed time microscopy allowed calculating the scoring time needed to reach certain accuracy in the aberration yield assessment, and from that, finding the optimal time schedule for the microscopy in different scenarios of exposure and workloads for the laboratory. It was found that in exposure scenarios leading to detrimental health effects (≥50% irradiation to ≥4 Gy), it is easier to reach a high number of Dic + CR than a high number cells scored; therefore, the former parameter can be recommended as more indicative for planning the work. In the cases of irradiations of ~2 Gy, ≥90% and of ~4 or ~12 Gy, ≥50%, which are health and life-threatening scenarios, the expertise level of biodosimetry can be provided with 5 h of microscopy that corresponds to the workload of 1 case per·day per·operator. Lethal situations of total-body doses of ~12 and ~20 Gy can be evaluated at the expertise level of accuracy with just 1 h of microscopy. By contrast to that, the scenario of a small fraction (10%) exposed to any dose within 2–12 Gy requires ≥25 h microscopy to score adequate numbers of Dic + CR.
For triage, it seems possible that scoring 25 Dic + CR is sufficient to maintain the relative error below 20% and that it can be achieved within 1 working day (5–6 h) by 1 operator for all scenarios, except the localized, 10% irradiations. The toughest situation occurs for a large-scale radiological event, in which the triage analysis scheme implies 10 cases per day. With 0.5 h of microscopy allowed per case and considering 10 Dic + CR as a tolerable threshold of minimum accuracy, only partial- and total-body (i.e., 90%–100%) exposure of 4–20 Gy can be identified. Meanwhile, scoring 50 cells for those scenarios would take ≥1 h; thus, microscopy time and not the number of scored metaphases should be recommended as a decision-making parameter for planning the triage biodosimetry.
One possible improvement of the cytogenetic procedures for increasing the accuracy of triage dose estimates is to use the QuickScan mode of analysis for faster dicentric acquisition.[1617] Another option is the use of motorized microscopes which allow hands-free operation for positioning the slide and focusing, avoiding the manual change of the objectives. However, automated dicentric analysis systems remain the ultimate solution for the triage biodosimetry. Due to technological advances over the last decade, such systems have become rather widespread across biodosimetry laboratories worldwide and there are a number of published reports on the evaluation of their practical effectiveness.[8911121314181920] It was shown that in 15 min, only approximately 10–15 cells can be counted manually while 20–25 can be counted with an automatic metaphase finder.[18] The main drawback of the automated systems is their high cost for the laboratories in low-income countries. Another limitation is related to the lower number of Dic that are identified during the automated scoring; however, this is partially compensated by the higher speed of metaphase finding. Sometimes, if only a few highly damaged but “scorable” cells are found by automated system, manual scoring is more time effective and therefore recommended.[14] In real-life situations, the discrepancy between the results of automated and manual scoring, particularly after partial-body exposures, may seriously affect the overall feasibility of interlaboratory networking and makes the usefulness of rapid, triage scoring in such exposure scenarios suspect.[19]
The data obtained in the present research can be used for a comparison of efficacy of the manual scoring against fully or partially automated analysis or motorized microscopy. Further investigations are needed to clarify the magnitude of possible inter-operator and inter-donor variability in “time versus accuracy” settings. This is very important in view of the standardization and networking within the biodosimetry service around the world. The datasets for such investigations have been already generated within interlaboratory comparisons based on sharing samples from many donors[1112] but still require proper statistical treatment.
The problems and questions, addressed in our study, are related not only to the conventional dicentric analysis but also to other two cytogenetic techniques suggested for emergency and triage biodosimetry – the premature chromosome condensation and micronuclei assays.[710212223] Therefore, our data and quantitative relationships linking the accuracy of the analysis, time of microscopy, cell scoring rate, and aberration yield in the sample can contribute to the improvement of the overall methodology of cytogenetic dosimetry. The suggested approach to generate a time scheme for provision of emergency/expertise biodosimetry service, depending on the workload and exposure scenario, might optimize a decision-making process in the management of the laboratory work in a large-scale radiological situation.
Conclusions
The research provided a set of experimental data, valuable for filling in gaps in the methodology of practical radiation cytogenetic biodosimetry. The microscopy time required for reaching a certain level of accuracy of chromosome damage measurement was assessed, and the accuracy achievable for certain times available for analysis was estimated in complex radiation exposure scenarios, simulated in vitro. The overall efficacy of chromosomal dosimetry, estimated as the scoring rate of either cells or aberrations, depends primarily on the aberration yield which is dependent on radiation dose and its distribution in a patient's body. For the first time, a strong negative correlation between the rate of aberration acquisition and the rate of cell recording was quantified. In practical settings, it makes it impossible to simultaneously acquire a high number of aberrations in a large sample of cells in a real-life biodosimetry service, which is usually limited in time. Therefore, the biodosimetry experts must always work in close contact with clinicians to decide what level of accuracy is preferable in each particular case.
The results of the exercise can be used to produce an optimal schedule of microscopy for different exposure scenarios, particularly involving high-dose homogeneous or partial-body irradiations. The workload for the biodosimetry laboratory of 1 case per operator per day (5 h of microscopy) allows dose estimates at the expertise level of accuracy for health and life-threatening situations, such as partial-body irradiations of 2 Gy or hemi-body irradiations of 4–12 Gy. Meanwhile, lethal total-body doses of 12 and 20 Gy can be evaluated at the expertise level with just 1 h of microscopy. In a large-scale radiological event, the triage analysis scheme of 0.5 h microscopy per case gives the minimum tolerable accuracy for distinguishing partial- and total-body exposure to doses from 4 to 20 Gy.
These simple algorithms and quantitative estimates can be instantly disseminated within the radiation cytogenetics community to be adopted by biodosimetry service laboratories and applied in practice, providing better health risk assessment and protection of the public in case of a radiological emergency.
Financial support and sponsorship
The work was carried out as a part of the project supported by the IAEA Research Contract 17079 in the framework of the IAEA Coordinated Research Project 3.50.08. This financial assistance was provided solely for author's activity in performing experimental studies reported in the present work. The IAEA did not influence or participate in study design; in the collection, analysis and interpretation of data; in the writing of the report; or in the decision to submit the paper for publication.
