- Record: found
- Abstract: found
- Article: not found
Designer Exosomes: A New Platform for Biotechnology Therapeutics

Read this article at
Abstract
Abstract
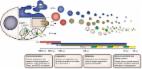
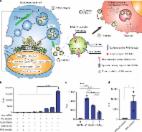
Desirable features of exosomes have made them a suitable manipulative platform for biomedical applications, including targeted drug delivery, gene therapy, cancer diagnosis and therapy, development of vaccines, and tissue regeneration. Although natural exosomes have various potentials, their clinical application is associated with some inherent limitations. Recently, these limitations inspired various attempts to engineer exosomes and develop designer exosomes. Mostly, designer exosomes are being developed to overcome the natural limitations of exosomes for targeted delivery of drugs and functional molecules to wounds, neurons, and the cardiovascular system for healing of damage. In this review, we summarize the possible improvements of natural exosomes by means of two main approaches: parental cell-based or pre-isolation exosome engineering and direct or post-isolation exosome engineering. Parental cell-based engineering methods use genetic engineering for loading of therapeutic molecules into the lumen or displaying them on the surface of exosomes. On the other hand, the post-isolation exosome engineering approach uses several chemical and mechanical methods including click chemistry, cloaking, bio-conjugation, sonication, extrusion, and electroporation. This review focuses on the latest research, mostly aimed at the development of designer exosomes using parental cell-based engineering and their application in cancer treatment and regenerative medicine.
Related collections
Most cited references92
- Record: found
- Abstract: found
- Article: found
Extracellular Vesicle Heterogeneity: Subpopulations, Isolation Techniques, and Diverse Functions in Cancer Progression
- Record: found
- Abstract: found
- Article: not found
Toward tailored exosomes: the exosomal tetraspanin web contributes to target cell selection.
- Record: found
- Abstract: found
- Article: found

