- Record: found
- Abstract: found
- Article: found
A Simple and Efficient Method for Visualizing Individual Cells in vivo by Cre-Mediated Single-Cell Labeling by Electroporation (CREMSCLE)
Read this article at
Abstract
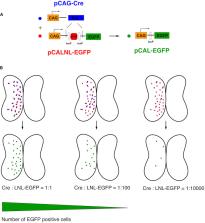
Efficient methods for visualizing cell morphology in the intact animal are of great benefit to the study of structural development in the nervous system. Quantitative analysis of the complex arborization patterns of brain cells informs cell-type classification, dissection of neuronal circuit wiring, and the elucidation of growth and plasticity mechanisms. Time-lapse single-cell morphological analysis requires labeling and imaging of single cells in situ without contamination from the ramified processes of other nearby cells. Here, using the Xenopus laevis optic tectum as a model system, we describe CRE-Mediated Single-Cell Labeling by Electroporation (CREMSCLE), a technique we developed based on bulk co-electroporation of Cre-dependent inducible expression vectors, together with very low concentrations of plasmid encoding Cre recombinase. This method offers efficient, sparse labeling in any brain area where bulk electroporation is possible. Unlike juxtacellular single-cell electroporation methods, CREMSCLE relies exclusively on the bulk electroporation technique, circumventing the need to precisely position a micropipette next to the target cell. Compared with viral transduction methods, it is fast and safe, generating high levels of expression within 24 h of introducing non-infectious plasmid DNA. In addition to increased efficiency of single-cell labeling, we confirm that CREMSCLE also allows for efficient co-expression of multiple gene products in the same cell. Furthermore, we demonstrate that this method is particularly well-suited for labeling immature neurons to follow their maturation over time. This approach therefore lends itself well to time-lapse morphological studies, particularly in the context of early neuronal development and under conditions that prevent more difficult visualized juxtacellular electroporation.
Related collections
Most cited references26
- Record: found
- Abstract: found
- Article: not found
Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral.
- Record: found
- Abstract: found
- Article: not found
Efficient in utero gene transfer system to the developing mouse brain using electroporation: visualization of neuronal migration in the developing cortex.
- Record: found
- Abstract: found
- Article: not found